Abstract
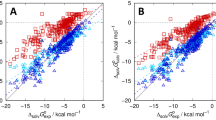
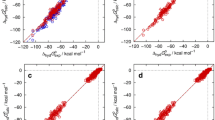
Implicit solvent methods for classical molecular modeling are frequently used to provide fast, physics-based hydration free energies of macromolecules. Less commonly considered is the transferability of these methods to other solvents. The Statistical Assessment of Modeling of Proteins and Ligands 5 (SAMPL5) distribution coefficient dataset and the accompanying explicit solvent partition coefficient reference calculations provide a direct test of solvent model transferability. Here we use the 3D reference interaction site model (3D-RISM) statistical-mechanical solvation theory, with a well tested water model and a new united atom cyclohexane model, to calculate partition coefficients for the SAMPL5 dataset. The cyclohexane model performed well in training and testing (\(R=0.98\) for amino acid neutral side chain analogues) but only if a parameterized solvation free energy correction was used. In contrast, the same protocol, using single solute conformations, performed poorly on the SAMPL5 dataset, obtaining \(R=0.73\) compared to the reference partition coefficients, likely due to the much larger solute sizes. Including solute conformational sampling through molecular dynamics coupled with 3D-RISM (MD/3D-RISM) improved agreement with the reference calculation to \(R=0.93\). Since our initial calculations only considered partition coefficients and not distribution coefficients, solute sampling provided little benefit comparing against experiment, where ionized and tautomer states are more important. Applying a simple \(\hbox {p}K_{\text {a}}\) correction improved agreement with experiment from \(R=0.54\) to \(R=0.66\), despite a small number of outliers. Better agreement is possible by accounting for tautomers and improving the ionization correction.








Similar content being viewed by others
References
Palmer DS, Frolov AI, Ratkova EL, Fedorov MV (2010) J Phys Condens Matter 22(49):492101. doi:10.1088/0953-8984/22/49/492101. http://stacks.iop.org/0953-8984/22/i=49/a=492101
Truchon JF, Pettitt BM, Labute P (2014) J Chem Theory Comput 10(3):934. doi:10.1021/ct4009359
Sergiievskyi V, Jeanmairet G, Levesque M, Borgis D (2015) J Chem Phys 143(18):184116. doi:10.1063/1.4935065. http://scitation.aip.org/content/aip/journal/jcp/143/18/10.1063/1.4935065
Misin M, Fedorov MV, Palmer DS (2015) J Chem Phys 142(9):091105. doi:10.1063/1.4914315. http://scitation.aip.org/content/aip/journal/jcp/142/9/10.1063/1.4914315
Misin M, Fedorov MV, Palmer DS (2016) J Phys Chem B 120(5):975. doi:10.1021/acs.jpcb.5b10809
Ratkova EL, Palmer DS, Fedorov MV (2015) Chem Rev 115(13):6312. doi:10.1021/cr5000283
Kovalenko A, Hirata F (2000) J Chem Phys 113(7):2793. doi:10.1063/1.1305885. http://scitation.aip.org/content/aip/journal/jcp/113/7/10.1063/1.1305885
Kido K, Yokogawa D, Sato H (2012) J Chem Phys 137(2):024106. doi:10.1063/1.4733393. http://scitation.aip.org/content/aip/journal/jcp/137/2/10.1063/1.4733393
Joung IS, Luchko T, Case DA (2013) J Chem Phys 138(4):044103. doi:10.1063/1.4775743. http://jcp.aip.org/resource/1/jcpsa6/v138/i4/p044103_s1
Kovalenko A, Hirata F (2000) J Chem Phys 112(23):10391. doi:10.1063/1.481676. http://scitation.aip.org/content/aip/journal/jcp/112/23/10.1063/1.481676
Kovalenko A, Hirata F (2000) J Chem Phys 112(23):10403. doi:10.1063/1.481677. http://scitation.aip.org/content/aip/journal/jcp/112/23/10.1063/1.481677
Johnson J, Case DA, Yamazaki T, Gusarov S, Kovalenko A, Luchko T (2016) J Phys Condens Matter 28(34):344002. doi:10.1088/0953-8984/28/34/344002. http://stacks.iop.org/0953-8984/28/i=34/a=344002
Ten-no S, Jung J, Chuman H, Kawashima Y (2010) Mol Phys 108(3–4):327. doi:10.1080/00268970903451848
Huang W, Blinov N, Kovalenko A (2015) J Phys Chem B 119(17):5588. doi:10.1021/acs.jpcb.5b01291
Misin M, Palmer DS, Fedorov MV (2016) J Phys Chem B 120(25):5724. doi:10.1021/acs.jpcb.6b05352
Bannan CC, Burley KH, Chiu M, Shirts MR, Gilson MK, Mobley DL (2016) J Comput Aided Mol Des. doi:10.1007/s10822-016-9954-8
Rustenburg AS, Justin Dancer BL, Ortwine DF, Mobley DL, Chodera JD (2016) J Comput Aided Mol Des (in press)
Tielker N, Tomazic D, Heil J, Ehrhart TKS, GÃŒssregen S, Schmidt KF, Kast SM (2016) J Comput Aided Mol Des (in press)
Luchko T, Gusarov S, Roe DR, Simmerling C, Case DA, Tuszynski J, Kovalenko A (2010) J Chem Theory Comput 6(3):607. doi:10.1021/ct900460m
Omelyan I, Kovalenko A (2015) J Chem Theory Comput 11(4):1875. doi:10.1021/ct5010438
Miyata T, Hirata F (2007) J Comput Chem 29:871
Omelyan I, Kovalenko A (2013) J Chem Phys 139(24):244106. doi:10.1063/1.4848716. http://scitation.aip.org/content/aip/journal/jcp/139/24/10.1063/1.4848716
Kovalenko A (2003) In: Hirata F (ed) Molecular theory of solvation, understanding chemical reactivity, vol 24. Kluwer Academic Publishers, Dordrecht, pp 169–275
Beglov D, Roux B (1997) J Phys Chem B 101(39):7821
Kovalenko A, Hirata F (1999) J Chem Phys 110(20):10095
Hansen JP, McDonald IR (1986) Theory of simple liquids. Academic Press, London
Hirata F (2003) In: Hirata F (ed) Molecular theory of solvation, understanding chemical reactivity, vol 24. Kluwer Academic Publishers, Dordrecht, pp 1–60
Perkyns JS, Pettitt BM (1992) Chem Phys Lett 190(6):626
Hirata F, Pettitt BM, Rossky PJ (1982) J Chem Phys 77(1):509. doi:10.1063/1.443606. http://scitation.aip.org/content/aip/journal/jcp/77/1/10.1063/1.443606
Hirata F, Rossky PJ, Pettitt BM (1983) J Chem Phys 78(6):4133. doi:10.1063/1.445090. http://scitation.aip.org/content/aip/journal/jcp/78/6/10.1063/1.445090
Kovalenko A (2013) Pure Appl Chem 85(1):159. doi:10.1351/PAC-CON-12-06-03
Yamazaki T, Blinov N, Wishart D, Kovalenko A (2008) Biophys J 95(10):4540. doi:10.1529/biophysj.107.123000. http://www.sciencedirect.com/science/article/pii/S0006349508785953
Imai T, Ohyama S, Kovalenko A, Hirata F (2007) Protein Sci 16(9):1927
Drabik P, Gusarov S, Kovalenko A (2007) Biophys J 92(2):394
Harano Y, Imai T, Kovalenko A, Kinoshita M, Hirata F (2001) J Chem Phys 114(21):9506
Imai T, Harano Y, Kinoshita M, Kovalenko A, Hirata F (2007) J Chem Phys 126(22):225102
Imai T, Harano Y, Kinoshita M, Kovalenko A, Hirata F (2006) J Chem Phys 125(2):024911
Imai T, Hiraoka R, Kovalenko A, Hirata F (2007) Proteins Struct Funct Bioinform 66(4):804
Imai T, Hiraoka R, Seto T, Kovalenko A, Hirata F (2007) J Phys Chem B 111(39):11585
Imai T, Isogai H, Seto T, Kovalenko A, Hirata F (2006) J Phys Chem B 110(24):12149
Imai T, Kovalenko A, Hirata F (2006) Mol Simul 32(10–11):817
Imai T, Hiraoka R, Kovalenko A, Hirata F (2005) J Am Chem Soc 127(44):15334
Huang W, Blinov N, Kovalenko A (2015) J Phys Chem B 119(17):5588. doi:10.1021/acs.jpcb.5b01291
Stumpe MC, Blinov N, Wishart D, Kovalenko A, Pande VS (2011) J Phys Chem B 115(2):319. doi:10.1021/jp102587q
Kaminski JW, Gusarov S, Wesolowski TA, Kovalenko A (2010) J Phys Chem A 114(20):6082. doi:10.1021/jp100158h
Kast SM, Kloss T (2008) J Chem Phys 129(23):236101. doi:10.1063/1.3041709. http://scitation.aip.org/content/aip/journal/jcp/129/23/10.1063/1.3041709
Imai T, Kinoshita M, Hirata F (2000) J Chem Phys 112(21):9469. doi:10.1063/1.481565. http://scitation.aip.org/content/aip/journal/jcp/112/21/10.1063/1.481565
Tuckerman ME, Berne BJ, Martyna GJ (1991) J Chem Phys 94(10):6811. doi:10.1063/1.460259. http://scitation.aip.org/content/aip/journal/jcp/94/10/10.1063/1.460259
Tuckerman M, Berne BJ, Martyna GJ (1992) J Chem Phys 97(3):1990. doi:10.1063/1.463137. http://scitation.aip.org/content/aip/journal/jcp/97/3/10.1063/1.463137
Ng K (1974) J Chem Phys 61(7):2680. doi:10.1063/1.1682399. http://scitation.aip.org/content/aip/journal/jcp/61/7/10.1063/1.1682399
Mobley DL, Dill KA, Chodera JD (2008) J Phys Chem B 112(3):938. doi:10.1021/jp0764384
Gilson MK, Given JA, Bush BL, McCammon JA (1997) Biophys J 72(3):1047. doi:10.1016/S0006-3495(97)78756-3. http://www.cell.com/article/S0006349597787563/abstract
Zwanzig RW (1954) J Chem Phys 22(8):1420. doi:10.1063/1.1740409. http://scitation.aip.org/content/aip/journal/jcp/22/8/10.1063/1.1740409
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) J Comput Chem 25(9):1157. doi:10.1002/jcc.20035. http://onlinelibrary.wiley.com/doi/10.1002/jcc.20035/abstract
Case DA, Babin V, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham TE, Darden TA, Duke RE, Gohlke H, Goetz AW, Gusarov S, Homeyer N, Janowski P, Kaus J, Kolossváry I, Kovalenko A, Lee TS, Le Grand S, Luchko T, Luo R, Madej B, Merz K, Paesani F, Roe DR, Roitberg AE, Sagui C, Salomon-Ferrer R, Seabra G, Simmerling C, Smith W, Swails JM, Walker RC, Wang J, Wolf RM, Wu X, Kollman PA (2015) AMBER 2015. University of California, San Francisco
Li J, Zhu T, Hawkins GD, Winget P, Liotard DA, Cramer CJ, Truhlar DG (1999) Theor Chem Acc 103(1):9. doi:10.1007/s002140050513. http://link.springer.com/article/10.1007/s002140050513
Radzicka A, Wolfenden R (1988) Biochemistry 27(5):1664. doi:10.1021/bi00405a042
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH (2016) Nucleic Acids Res 44(D1):D1202. doi:10.1093/nar/gkv951. http://nar.oxfordjournals.org/content/44/D1/D1202
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) J Cheminform 3(1):33. doi:10.1186/1758-2946-3-33. http://www.jcheminf.com/content/3/1/33/abstract
National Center for Biotechnology Information. PubChem Compound Database; CID=8078. https://pubchem.ncbi.nlm.nih.gov/compound/8078. Accessed 14 Jan 2016
Perkyns J, Pettitt BM (1992) J Chem Phys 97(10):7656. doi:10.1063/1.463485. http://scitation.aip.org/content/aip/journal/jcp/97/10/10.1063/1.463485
Hirata F, Rossky PJ (1981) Chem Phys Lett 83(2):329. doi:10.1016/0009-2614(81)85474-7. http://www.sciencedirect.com/science/article/pii/0009261481854747
Yang L, Tan Ch, Hsieh MJ, Wang J, Duan Y, Cieplak P, Caldwell J, Kollman PA, Luo R (2006) J Phys Chem B 110(26):13166. doi:10.1021/jp060163v. http://pubs.acs.org/doi/abs/10.1021/jp060163v
Schuler LD, Daura X, van Gunsteren WF (2001) J Comput Chem 22(11):1205. doi:10.1002/jcc.1078. http://onlinelibrary.wiley.com/doi/10.1002/jcc.1078/abstract
Chandler D, Andersen HC (1972) J Chem Phys 57(5):1930. doi:10.1063/1.1678513. http://scitation.aip.org/content/aip/journal/jcp/57/5/10.1063/1.1678513
Aicart E, Tardajos G, Diaz Pena M (1980) J Chem Eng Data 25(2), 140. doi:10.1021/je60085a007. http://dx.doi.org/10.1021/je60085a007
Anikeenko AV, Kim AV, Medvedev NN (2010) J Struct Chem 51(6), 1090. http://link.springer.com/article/10.1007/s10947-010-0167-z
Shumway RH, Stoffer DS (2010) Time series analysis and its applications: with R examples. Springer, Berlin
chemicalize.org was used for prediction of acid and base pk\(_a\) values (2016) http://www.chemaxon.com. Chemicalize.org and ChemAxon
Manchester J, Walkup G, Rivin O, You Z (2010) J Chem Inf Model 50(4):565. doi:10.1021/ci100019p
MacCallum JL, Tieleman DP (2003) J Comput Chem 24(15):1930. doi:10.1002/jcc.10328. http://onlinelibrary.wiley.com/doi/10.1002/jcc.10328/abstract
Villa A, Mark AE (2002) J Comput Chem 23(5):548. doi:10.1002/jcc.10052. http://onlinelibrary.wiley.com/doi/10.1002/jcc.10052/abstract
Chang J, Lenhoff AM, Sandler SI (2007) J Phys Chem B 111(8):2098. doi:10.1021/jp0620163
Oostenbrink C, Villa A, Mark AE, Van Gunsteren WF (2004) J Comput Chem 25(13):1656. doi:10.1002/jcc.20090. http://onlinelibrary.wiley.com/doi/10.1002/jcc.20090/abstract
Acknowledgments
The authors would like to thank Caitlin C. Bannan and David L. Mobley for access to the results from their explicit solvent reference calculations. T.L. would like to additionally thank David Mobley and Stefan Kast for useful discussions about calculating solvation free energies from time series. This work was partially supported by the California State University Program for Education and Research in Biotechnology (CSUPERB; T.L., G.C.L., K.P.J.) and by the Alberta Prion Research Institute and the National Research Council of Canada (N.B., A.K.).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10822_2016_9947_MOESM1_ESM.pdf
The online version of this article contains supplementary material, including discussion of conformational sampling issues, partition coefficients for neutral amino acid side chains, solvation free energies computed by alternate thermodynamic paths, and \(\hbox {p}K_{\text {a}}\) predictions.
Rights and permissions
About this article
Cite this article
Luchko, T., Blinov, N., Limon, G.C. et al. SAMPL5: 3D-RISM partition coefficient calculations with partial molar volume corrections and solute conformational sampling. J Comput Aided Mol Des 30, 1115–1127 (2016). https://doi.org/10.1007/s10822-016-9947-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-016-9947-7