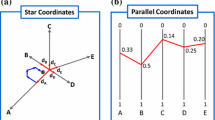
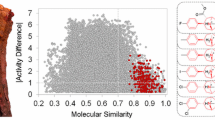
Abstract
Activity landscapes (ALs) of compound data sets are rationalized as graphical representations that integrate similarity and potency relationships between active compounds. ALs enable the visualization of structure–activity relationship (SAR) information and are thus computational tools of interest for medicinal chemistry. For AL generation, similarity and potency relationships are typically evaluated in a pairwise manner and major AL features are assessed at the level of compound pairs. In this study, we add a conditional probability formalism to AL design that makes it possible to quantify the probability of individual compounds to contribute to characteristic AL features. Making this information graphically accessible in a molecular network-based AL representation is shown to further increase AL information content and helps to quickly focus on SAR-informative compound subsets. This feature probability-based AL variant extends the current spectrum of AL representations for medicinal chemistry applications.




Similar content being viewed by others
References
Wassermann AM, Wawer M, Bajorath J (2010) Activity landscape representations for structure–activity relationship analysis. J Med Chem 53(23):8209–8223
Stumpfe D, Bajorath J (2012) Methods for SAR visualization. RSC Adv 2(2):369–378
Iyer P, Stumpfe D, Vogt M, Bajorath J, Maggiora GM (2013) Activity landscapes, information theory, and structure–activity relationships. Mol Inf 32(5–6):421–430
Wawer M, Peltason L, Weskamp N, Tecketrup A, Bajorath J (2008) Structure–activity relationship anatomy by network-like similarity graphs and local structure–activity relationship indices. J Med Chem 51(19):6075–6084
Peltason L, Iyer P, Bajorath J (2010) Rationalizing three-dimensional activity landscapes and the influence of molecular representations on landscape topology and the formation of activity cliffs. J Chem Inf Model 50(6):1021–1033
Stumpfe D, Bajorath J (2012) Exploring activity cliffs in medicinal chemistry. J Med Chem 55(7):2932–2942
Stumpfe D, Hu Y, Dimova D, Bajorath J (2014) Recent progress in understanding activity cliffs and their utility in medicinal chemistry. J Med Chem 57(1):18–28
Vogt M, Iyer P, Maggiora GM, Bajorath J (2013) Conditional probabilities of activity landscape features for individual compounds. J Chem Inf Model 53(7):1602–1612
Willett P, Barnard JM, Downs GM (1998) Chemical similarity searching. J Chem Inf Comput Sci 38(6):983–996
Rogers D, Hahn M (2010) Extended-connectivity fingerprints. J Chem Inf Model 50(5):742–754
Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 40(Database issue):D1100–D1107
OEChem TKv 2013 April (2013) OpenEye Scientific Software Inc, Santa Fe, New Mexico
Acknowledgments
The authors thank Dr. Dilyana Dimova for providing activity landscape models using alternative similarity measures and Dr. Ye Hu for help with data sets. B.Z. is supported by the China Scholarship Council.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, B., Vogt, M. & Bajorath, J. Design of an activity landscape view taking compound-based feature probabilities into account. J Comput Aided Mol Des 28, 919–926 (2014). https://doi.org/10.1007/s10822-014-9773-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-014-9773-8