Summary
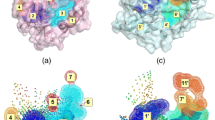
This paper presents a new algorithm to compare substructural epitopes in protein binding cavities. Through the comparison of binding cavities accommodating well characterized ligands with cavities whose actual guests are yet unknown, it is possible to draw some conclusions on the required shape of a putative ligand likely to bind to the latter cavities. To detect functional relationships among proteins, their binding-site exposed physicochemical characteristics are described by assigning generic pseudocenters to the functional groups of the amino acids flanking the particular active site. The cavities are divided into small local regions of four pseudocenters having the shape of a pyramid with triangular basis. To find similar local regions, an emergent self-organizing map is used for clustering. Two local regions within the same cluster are similar and form the basis for the superpositioning of the corresponding cavities to score this match. First results show that the similarities between enzymes with the same EC number can be found correctly. Enzymes with different EC numbers are detected to have no common substructures. These results indicate the benefit of this method and motivate further studies.
Similar content being viewed by others
References
H.M. Grindley P.J. Artymiuk D.W. Rice P. Willett (1993) J. Mol. Biol., 229 707
R.V. Spriggs P.J. Artymiuk P. Willett (2003) J. Chem. Inf. Comput. Sci., 43 412
A.C. Wallace R.A. Laskowski J.M. Thornton (1996) Protein Sci., 5 1001
A.C. Wallace N. Borkakoti J.M. Thornton (1997) Protein Sci., 6 2308
R.B. Russell (1998) J. Mol. Biol., 279 1211
A. Stark S. Sunyaev R.B. Russell (2003) J. Mol. Biol., 326 1307
A. Stark R.B. Russell (2003) Nucleic Acids Res., 31 3341
P.P. Wangikar A.V. Tendulkar S. Ramya D.N. Mali S. Sarawagi (2003) J. Mol. Biol., 326 955
T. Hamelryck (2003) Proteins, 51 96
G.J. Kleywegt (1999) J. Mol. Biol., 285 1887
O. Bachar D. Fischer R. Nussinov H. Wolfson (1993) Protein Eng., 6 279
M. Rosen S.L. Lin H. Wolfson R. Nussinov (1998) Protein Eng., 11 263
K. Kinoshita H. Nakamura (2003) Protein Sci., 12 1589
J.V. Lehtonen K. Denessiouk A.C. May M.S. Johnson (1999) Proteins, 34 341
A.R. Poirrette P.J. Artymiuk D.W. Rice P. Willett (1997) J. Comput.-Aided Mol. Des., 11 557
A. Yan J. Gasteiger (2003) J. Chem. Inf. Comput. Sci., 43 429
M. Stahl C. Taroni G. Schneider (2000) Protein Eng. 13 IssueID2 83
S. Schmitt D. Kuhn G. Klebe (2002) J. Mol. Biol., 323 387
T. Kohonen (1982) Biol. Cybernetics, 43 59
Ultsch, A., Proc. Workshop on Self-organizing Maps, Kyushu, Japan, 2003, pp. 225–230.
Ultsch, A., Proc. Conf. Soc. for Information and Classification, 1992.
A. Ultsch (2004) U-Matrix a tool to visualize clusters in high dimensional data. Technical Report No. 36 Department of Mathematics and Computer Science University of Marburg
M. Hendlich F. Rippmann G. Barnickel (1997) J. Mol. Graph. Model, 15 359
M. Hendlich A. Bergner J. Gunther G. Klebe (2003) J. Mol. Biol. 326 607
A. Bairoch (2000) Nucleic Acids Res. 28 IssueID1 304
Siemon, R., Einige Werkzeuge zum Einsatz von selbstorganisierenden Neuronalen Netzen zur Strukturanalyse von Wirkstoff-Rezeptoren. Diploma Thesis, Department of Mathematics and Computer Science, University of Marburg, 2001.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kupas, K., Ultsch, A. & Klebe, G. Comparison of substructural epitopes in enzyme active sites using self-organizing maps. J Comput Aided Mol Des 18, 697–708 (2004). https://doi.org/10.1007/s10822-004-6553-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-004-6553-x