Abstract
Purpose
Macrozoospermia is a rare sperm morphologic abnormality associated with male infertility and is characterized by a high percentage of spermatozoa with large irregular heads. The aim of this study was to identify the genetic cause of an infertile male with macrozoospermia from a consanguineous family.
Methods
Whole-exome sequencing (WES) was performed using peripheral blood genomic DNA from the patient and his parents.
Results
WES analysis of the patient with macrozoospermia from a consanguineous family allowed the identification of a novel homozygous missense variant in the AURKC gene (c.269G>A). Bioinformatics analysis also suggested this variant a pathogenic mutation. Quantitative real-time PCR analysis showed that the mRNA level of AURKC is significantly decreased in the patient compared with his father. Moreover, no embryos were available for transfer after ICSI.
Conclusions
These results further support the important role of AURKC in male infertility and guide the practitioner in optimal decision making for patients with macrozoospermia.
Similar content being viewed by others
Introduction
Approximately 70 million couples worldwide suffer from infertility [1], and approximately half of these cases are due to male factors [2]. Macrozoospermia is a rare sperm morphologic abnormality associated with male infertility and is characterized by a high percentage of spermatozoa with large irregular heads [3, 4]. This syndrome, first reported in 1977, affects < 1% of infertile men [3]. It is considered to be an autosomal recessive type of teratozoospermia that results in male infertility [5]. Currently, the most relevant single-gene defect that has been identified in a patient with macrozoospermia is a mutation in Aurora Kinase C (AURKC) [5,6,7].
The AURKC gene encodes a member of a highly conserved serine/threonine kinase family, which plays crucial roles in centrosome function, homologous chromosome segregation, and cytokinesis during meiosis [7, 8]. Until now, only five mutations of AURKC have been described to be associated with macrozoospermia: c.144delC (p.L499Wfs22), c.744C>G (p.Y248*), c686G>A (p.C229Y), c.930+38G>A (occurs in the 3′-UTR), and c.436-2A>G (splicing site mutation that leads to the skipping of exons 5) [6,7,8,9,10,11,12,13]. These mutations in humans lead to reduced protein function, which results in meiotic failure, but spermatogenesis is not affected, leading to the production of large-headed spermatozoa [9, 10, 14].
Here, we report a novel homozygous missense variant in the AURKC gene in an infertile male from a consanguineous family identified via whole-exome sequencing. In addition, this variant has a high probability of pathogenicity according to in silico analysis. ICSI was also performed and failed to generate any embryos suitable for transfer. These results further support the important role of AURKC in male infertility and guide the practitioner in optimal decision-making for patients with macrozoospermia.
Materials and methods
Patient
The proband was a 27-year-old treated for infertility at Anhui Provincial Hospital. The results of the patient’s semen tests are described in Table 1. The results revealed that the patient suffered from macrozoospermia (close to 100% large-headed spermatozoa with a sperm count of 1 M/ml) (Table 1). The parents of the patient were first-degree cousins (Fig. 1). The patient exhibited normal erection and ejaculation, and reported having regular sexual intercourse 2–3 times/week; however, his wife had been unable to get pregnant since they were married in 2015. The patient had no history of unhealthy activity or contact with adverse chemicals. Physical examination revealed normally developed male external genitalia, normal bilateral testicular size, and no abnormality in the bilateral spermatic veins upon palpation. The proband did not have primary microcephaly or respiratory disease. The chromosomal karyotype of the patient was normal (46; XY), and no deletion was found in the Y chromosome. The patient hormone levels were normal.
This project was approved by the ethics committee of Anhui Medical University. All study members gave their written informed consent before sample collection.
Genomic DNA extraction and whole-exome sequencing (WES)
The genomic DNA from the patient and his parents was extracted from peripheral blood samples using a QIAamp DNA blood midi kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol.
Genomic DNA from the proband and his parents was subjected to WES. WES was performed by the WuXi NextCODE in Shanghai on a HiSeq2000 sequencing platform (Illumina, San Diego, CA, USA). After the adaptors were removed, the WES raw reads were aligned to the reference genome Hg19 using the Burrows-Wheeler aligner, followed by removal of PCR duplicates. Variants including single-nucleotide polymorphisms and indels were identified using SAMtools and annotated by ANNOVAR software. A candidate gene was considered a variant that fulfilled the following criteria: (i) missense, nonsense, frame shift, and splice site variants, (ii) absent or rare (frequency below 1%) in the two databases (dbSNP, 1000G), and (iii) homozygous variants in the patient and heterozygous variants in his parents.
Sanger sequencing validation
The mutation in AURKC in the proband and his parents was validated using Sanger sequencing. We amplified the PCR products of exon 3 of the AURKC gene using specific primes (the forward primer is 5′-AACCAGGATTCGAGTGTCTG-3′, and the reverse primer is 5′-CAATCTCCAGGTAGACGATGGAG-3′). Then, the PCR products were sequenced on an ABI 3730XL automated sequencer (Applied Biosystems, Forster City, CA, USA).
RNA extraction and Q-PCR
RNA extraction was carried out on the whole blood using TRI REAGENT®BD (Molecular Research Center) using the manufacturer’s protocol. Reverse transcription was performed in patient and his parents with 5 μl of extracted RNA. Hybridization of the oligo-dT was performed by incubating for 30 min at 42 °C and quenching on ice with the following mix: 5 μl of RNA, 10 μl of 2Xsupermix (10 mM, Pharmacia), 1 μl of gDNA (0.5 mM, Roche diagnostics), and 4 μl of H2O. Then 2 μl of the obtained cDNA mix was used for the quantitative PCR (Q-PCR) using a StepOne-PlusTM Real-Time PCR System (Life Technologies) with Power SYBR Green PCR Master Mix (Life Technologies) according to the manufacturer’s protocol. Quantification of the fold change in gene expression was determined by the relative quantification method (2−ΔΔCT) using the gapdh gene as a reference. Data are shown as the average fold increase standard error of the mean. Primers are described in supplementary file 1.
ICSI, embryo, and evaluation of embryo quality
The patient and his wife underwent intracytoplasmic sperm injection (ICSI) at Anhui Provincial Hospital. Briefly, embryo culture was performed using Vitrolife G-SERIES™ culture media (Vitrolife, Goteborg, Sweden) according to the manufacturer’s instructions. The patient’s wife had undergone one ICSI cycle. Detailed results are described in the “Results” section.
Sequence alignment of AURKC protein
Sequence alignment of the AURKC protein in different species was conducted using ClustalX2.1. The number of each species was as follows: Homo sapiens (NP_001015878.1), Mus musculus (AAI00338.1), Bos Taurus (NP_001180124.1), Desmodus rotundus (XP_024425801.1), Pan paniscus (XP_003816716.1), Pongo abelii (XP_002829903.1), and Rattus norvegicus (NP_001295465.1).
Results
WES analysis of a patient with macrozoospermia
To identify the genetic cause of the macrozoospermia, we performed WES using peripheral blood genomic DNA from the patient and his parents to identify the putative pathogenic mutation. Considering the family history of consanguinity, we focused on homozygous mutations. After of the exclusions of frequent variants and application of technical and biological filters (see “Materials and methods” section and Fig. 2A), a limited list of homozygous mutations was established (Supplementary Table 1). To determine whether any of these 36 genes may be related to macrozoospermia, we first examined the tissue expression patterns for each gene. Among these homozygous variant genes, only two genes exhibited testis-enriched expression (Fig. S1). One of the identified genes was AURKC, and a new mutation (c.269G>A, GenBank accession number, NM_001015878) in exon 3 was identified in the patient by WES. Accordingly, the change in amino acids was determined to be Arg90Gln. Another identified gene was gametogenetin (GGN), and a mutation (c.148T>C, GenBank accession number, NM_152657.3) in exon 3 was identified in the patient by WES. Accordingly, the change in amino acids was determined to be Trp50Arg.
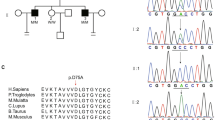
Whole-exome sequencing analysis of the consanguineous family. A Filter strategies used in this study. B Sanger sequencing validation of the affected patient and his parents. The arrow indicates the mutation site. CQ-PCR analysis of mRNA expressions of AURKC in the patient and his father. Messenger RNA expression determined by Q-PCR is calculated as a ratio relative to gapdh and expressed relative to his father. D Sequence alignment of AURKC protein in different species. The arrow refers to the mutation site
Validation by Sanger sequencing
We next performed Sanger sequencing to verify the homozygous variant in the AURKC and GGN genes in both the patient and his parents. Exon 3 of the AURKC gene was amplified from genomic DNA of the patient and his parents using PCR. The homozygous missense variant in AURKC was validated in the patient, and his parents had heterozygous alleles (Fig. 2B).
Detrimental effects of the identified variant in AURKC
Unfortunately, we could not get access to testicular biopsy samples from the affected patient and his parents; therefore, we set up Q-PCR to study the effect of the variation. Q-PCR analysis demonstrated that mRNA levels of AURKC were down-regulated significantly in the patient (Fig. 2C).
In silico analysis of the mutation
Bioinformatics analysis of the c.269G>A mutation in the AURKC gene by three online pathogenicity prediction tools (Polyphen-2, SIFT, Mutation taster) suggested that this mutation is most likely a disease-causing mutation (Supplementary Table 2). This variant is a novel mutation that was absent in the ExAC, 1000G, and gnomAD databases. The mutation site at R90 was highly conserved from human to zebrafish, indicating an important role of this site for the function of AURKC protein (Fig. 2D).
ICSI using the patient’s sperm
After centrifugation on density gradients and careful examination, a few “normal-looking” spermatozoa that fit into an ICSI micropipette were selected. One ICSI cycle was attempted for the patient at Anhui Provincial Hospital. For the ICSI cycles, we collected 10 eggs and 7 MII oocytes; none of the embryos developed to the blastocyst stage.
Discussion
In the last decade, five mutations (c.144delC (p.L49Wfs22), c.744C>G (p.Y248*), c686G>A (p.C229Y), c.930+38G>A (occurs in the 3′-UTR), and c.436-2A>G (splicing site mutation that leads to the skipping of exons 5)) [8,9,10,11,12,13] in the AURKC gene have been reported to be associated with macrozoospermia. In this study, we identified a novel homozygous mutation (c.269 G>A; p.R90Q) in the AURKC gene in a macrozoocephalic patient from a consanguineous family. One ICSI cycle was carried out in the patient, but none of the embryos developed to the blastocyst stage.
To date, numerous reports have provided evidence suggesting that failures in chromosome segregation and/or cytokinesis during the first, second, or both meiotic divisions are the major cause of macrozoospermia [15,16,17,18,19]. Aurora kinases are highly evolutionarily conserved kinases that are required for accurate chromosome segregation and cytokinesis during meiosis [20]. AURKC has 7 exons and encodes a 309 amino acid protein in humans. AURKC is a component of the Aurora kinase family and is predominantly expressed in the testis [21, 22]. In this study, we found a new missense mutation in exon 3 of the AURKC gene that led to an amino acid change (p.R90Q). The sequence alignment of the AURKC protein showed that this mutation site is conserved among different species (Fig. 2C). We utilized three online pathogenicity predict tools (Polyphen-2, SIFT, Mutation taster) to predict the harmfulness of this variant (Table 2). The results suggested that this variant has a high probability of pathogenicity.
Previous studies have compared the values of a routine spermogram and spermocytogram between patients with and without an AURKC mutation [8]. The results showed that the proportions of large-headed spermatozoa generally reached far higher values, and the presence of 1% normal spermatozoa is the most discriminant parameter in patients with AURKC mutations [8]. Consistent with previous studies, the patient with missense variant in the AURKC gene (c.269G>A) in this study showed 96% large-headed spermatozoa. However, the proportions of normal spermatozoa reached 4%. One explanation could be that in addition to macrozoospermia, this patient studied here also has a low sperm number (1.71 × 106). Variable scoring of the normal spermatozoa features between operators and laboratories could also influence the results as some spermatozoa with a slight morphology defect in head or flagella could be considered normal [8].
Numerous publications have described a failure of pregnancy in patients with macrozoospermia [5, 7, 12, 16,17,18, 23,24,25]. In addition, further studies have recommended that systematic genetic screening of the AURKC gene be performed when a patient has more than 30% enlarged head spermatozoa. If a mutation in the AURKC gene is found, ICSI should not be attempted [7, 26]. In this study, the patient’s wife failed to become pregnant after one ICSI cycle, again demonstrating the correlation between AURKC gene mutation and ICSI outcome. Consequently, ICSI in such a patient is not recommended.
In conclusion, we identified a novel missense mutation (c.269 G>A; p.R90Q) in the AURKC gene. To date, this is the sixth reported variant in the AURKC gene associated with macrozoospermia. This study expands the spectrum of AURKC mutations and helps guide the practitioners to make optimal decisions for patients with macrozoospermia.
References
Boivin J, Bunting L, Collins JA, Nygren KG. International estimates of infertility prevalence and treatment-seeking: potential need and demand for infertility medical care. Hum Reprod. 2007;22(6):1506–12.
Silber SJ. Evaluation and treatment of male infertility. Clin Obstet Gynecol. 2000;43(4):854–88.
Nistal M, Paniagua R, Herruzo A. Multi-tailed spermatozoa in a case with asthenospermia and teratospermia. Virchows Arch B Cell Pathol. 1977;26(2):111–8.
Brahem S, Mehdi M, Elghezal H, Saad A. Study of aneuploidy rate and sperm DNA fragmentation in large-headed, multiple-tailed spermatozoa. Andrologia. 2012;44(2):130–5.
Ghedir H, Gribaa M, Mamai O, Ben Charfeddine I, Braham A, Amara A, et al. Macrozoospermia: screening for the homozygous c.144delC mutation in AURKC gene in infertile men and estimation of its heterozygosity frequency in the Tunisian population. J Assist Reprod Genet. 2015;32(11):1651–8.
Dieterich K, Soto Rifo R, Faure AK, Hennebicq S, Ben Amar B, Zahi M, et al. Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat Genet. 2007;39(5):661–5.
Ben Khelifa M, Zouari R, Harbuz R, Halouani L, Arnoult C, Lunardi J, et al. A new AURKC mutation causing macrozoospermia: implications for human spermatogenesis and clinical diagnosis. Mol Hum Reprod. 2011;17(12):762–8.
Ben Khelifa M, Coutton C, Blum MG, Abada F, Harbuz R, Zouari R, et al. Identification of a new recurrent aurora kinase C mutation in both European and African men with macrozoospermia. Hum Reprod. 2012;27(11):3337–46.
Dieterich K, Zouari R, Harbuz R, Vialard F, Martinez D, Bellayou H, et al. The Aurora Kinase C c.144delC mutation causes meiosis I arrest in men and is frequent in the North African population. Hum Mol Genet. 2009;18(7):1301–9.
Harbuz R, Zouari R, Dieterich K, Nikas Y, Lunardi J, Hennebicq S, et al. Function of aurora kinase C (AURKC) in human reproduction. Gynecol Obstet Fertil. 2009;37(6):546–51.
El Kerch F, Lamzouri A, Laarabi FZ, Zahi M, Ben Amar B, Sefiani A. Confirmation of the high prevalence in Morocco of the homozygous mutation c.144delC in the aurora kinase C gene (AURKC) in the teratozoospermia with large-headed spermatozoa. J Gynecol Obstet Biol Reprod. 2011;40(4):329–33.
Ounis L, Zoghmar A, Coutton C, Rouabah L, Hachemi M, Martinez D, et al. Mutations of the aurora kinase C gene causing macrozoospermia are the most frequent genetic cause of male infertility in Algerian men. Asian J Androl. 2015;17(1):68–73.
Eloualid A, Rouba H, Rhaissi H, Barakat A, Louanjli N, Bashamboo A, et al. Prevalence of the Aurora kinase C c.144delC mutation in infertile Moroccan men. Fertil Steril. 2014;101(4):1086–90.
Fellmeth JE, Ghanaim EM, Schindler K. Characterization of macrozoospermia-associated AURKC mutations in a mammalian meiotic system. Hum Mol Genet. 2016;25(13):2698–711.
Escalier D, Bermudez D, Gallo JM, Viellefond A, Schrevel J. Cytoplasmic events in human meiotic arrest as revealed by immunolabelling of spermatocyte proacrosin. Differentiation. 1992;51(3):233–43.
In't Veld PA, Broekmans FJ, de France HF, Pearson PL, Pieters MH, van Kooij RJ. Intracytoplasmic sperm injection (ICSI) and chromosomally abnormal spermatozoa. Hum Reprod. 1997;12(4):752–4.
Weissenberg R, Aviram A, Golan R, Lewin LM, Levron J, Madgar I, et al. Concurrent use of flow cytometry and fluorescence in-situ hybridization techniques for detecting faulty meiosis in a human sperm sample. Mol Hum Reprod. 1998;4(1):61–6.
Benzacken B, Gavelle FM, Martin-Pont B, Dupuy O, Lievre N, Hugues JN, et al. Familial sperm polyploidy induced by genetic spermatogenesis failure: case report. Hum Reprod. 2001;16(12):2646–51.
Devillard F, Metzler-Guillemain C, Pelletier R, DeRobertis C, Bergues U, Hennebicq S, et al. Polyploidy in large-headed sperm: FISH study of three cases. Hum Reprod. 2002;17(5):1292–8.
Yang KT, Tang CJ, Tang TK. Possible role of Aurora-C in meiosis. Front Oncol. 2015;5:178.
Bernard M, Sanseau P, Henry C, Couturier A, Prigent C. Cloning of STK13, a third human protein kinase related to Drosophila aurora and budding yeast Ipl1 that maps on chromosome 19q13.3-ter. Genomics. 1998;53(3):406–9.
Tang CJ, Lin CY, Tang TK. Dynamic localization and functional implications of Aurora-C kinase during male mouse meiosis. Dev Biol. 2006;290(2):398–410.
Yurov YB, Saias MJ, Vorsanova SG, Erny R, Soloviev IV, Sharonin VO, et al. Rapid chromosomal analysis of germ-line cells by FISH: an investigation of an infertile male with large-headed spermatozoa. Mol Hum Reprod. 1996;2(9):665–8.
Lewis-Jones I, Aziz N, Seshadri S, Douglas A, Howard P. Sperm chromosomal abnormalities are linked to sperm morphologic deformities. Fertil Steril. 2003;79(1):212–5.
Perrin A, Morel F, Moy L, Colleu D, Amice V, De Braekeleer M. Study of aneuploidy in large-headed, multiple-tailed spermatozoa: case report and review of the literature. Fertil Steril. 2008;90(4):1201 e1213–07.
Carmignac V, Dupont JM, Fierro RC, Barberet J, Bruno C, Lieury N, et al. Diagnostic genetic screening for assisted reproductive technologies patients with macrozoospermia. Andrology. 2017;5(2):370–80.
Acknowledgements
We thank all the study participants.
Funding
This work was supported by the Natural Science Foundation of Anhui Provincial of China (1608085QH174).
Author information
Authors and Affiliations
Contributions
J.H. performed the majority of experiments. J.H. analyzed the WES data. J.H. wrote the paper. Peripheral blood samples were collected by YY.W. ICSI was done by YY.W. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
This project was approved by the ethics committee on human research of Anhui provincial hospital (Approve ID: 20131357).
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hua, J., Wan, Yy. Whole-exome sequencing identified a novel mutation of AURKC in a Chinese family with macrozoospermia. J Assist Reprod Genet 36, 529–534 (2019). https://doi.org/10.1007/s10815-018-1374-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-018-1374-3