Abstract
Purpose
To study the chromosome constitution of equal-sized three-cell embryo.
Methods
We determined the chromosome constitution of 105 blastomeres from 35 embryos using multiple annealing and looping-based amplification cycles (MALBAC) together with NGS sequencing technology. Chromosomal copy number variation (CNV) analysis was successfully performed in 27 embryos. We also analyzed radius, perimeter, area, and volume of each blastomere to explore the possibility of selecting the normal embryos.
Results
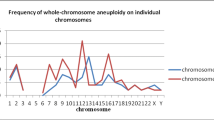
Majority of the embryos (77.8%, 21/27) studied were mosaic or aneuploid, and only 22.2% (6/27) had normal chromosome numbers. The aneuploid chromosomes spread across all chromosomes and the most frequent aneuploidies were for chromosomes 1, 16, and 18 followed by 13, 19, and 21. Statistical analyses showed no significant difference between euploid and aneuploid embryos regarding radius, perimeter, area, and volume of their blastomeres.
Conclusions
Our results showed that majority of the equal-sized three-cell embryos were chromosomally abnormal and could not be distinguished by morphology observation, so they should be given lower priority at selection for transfer.

Similar content being viewed by others
References
Alpha Scientists in Reproductive M, Embryology ESIGo. The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod. 2011;26:1270–83.
Prados FJ, Debrock S, Lemmen JG, Agerholm I. The cleavage stage embryo. Hum Reprod. 2012;27(Suppl 1):i50–71.
Hlinka D, Kalatova B, Uhrinova I, Dolinska S, Rutarova J, Rezacova J, et al. Time-lapse cleavage rating predicts human embryo viability. Physiol Res. 2012;61:513–25.
Athayde Wirka K, Chen AA, Conaghan J, Ivani K, Gvakharia M, Behr B, et al. Atypical embryo phenotypes identified by time-lapse microscopy: high prevalence and association with embryo development. Fertil Steril. 2014;101:1637–48 e1–5.
Telentschak S, Soliwoda M, Nohroudi K, Addicks K, Klinz FJ. Cytokinesis failure and successful multipolar mitoses drive aneuploidy in glioblastoma cells. Oncol Rep. 2015;33:2001–8.
Kalatova B, Jesenska R, Hlinka D, Dudas M. Tripolar mitosis in human cells and embryos: occurrence, pathophysiology and medical implications. Acta Histochem. 2015;117:111–25.
Chamayou S, Patrizio P, Storaci G, Tomaselli V, Alecci C, Ragolia C, et al. The use of morphokinetic parameters to select all embryos with full capacity to implant. J Assist Reprod Genet. 2013;30:703–10.
Zhan Q, Ye Z, Clarke R, Rosenwaks Z, Zaninovic N. Direct unequal cleavages: embryo developmental competence, genetic constitution and clinical outcome. PLoS One. 2016;11:e0166398.
Hardarson T, Selleskog U, Reismer E, Wigander A, Wennerstrom S, Westin C, et al. Zygotes cleaving directly into more than two cells after 25–27 h in culture are predominantly chromosomally abnormal. Hum Reprod. 2006;21:1.
Fiorentino F, Bono S, Biricik A, Nuccitelli A, Cotroneo E, Cottone G, et al. Application of next-generation sequencing technology for comprehensive aneuploidy screening of blastocysts in clinical preimplantation genetic screening cycles. Hum Reprod. 2014;29:2802–13.
Yan L, Huang L, Xu L, Huang J, Ma F, Zhu X, et al. Live births after simultaneous avoidance of monogenic diseases and chromosome abnormality by next-generation sequencing with linkage analyses. Proc Natl Acad Sci U S A. 2015;112:15964–9.
Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–83.
Lu Y, Peng H, Jin Z, Cheng J, Wang S, Ma M, et al. Preimplantation genetic diagnosis for a Chinese family with autosomal recessive Meckel-Gruber syndrome type 3 (MKS3). PLoS One. 2013;8:e73245.
Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–6.
Chan KC, Jiang P, Chan CW, Sun K, Wong J, Hui EP, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A. 2013;110:18761–8.
Kola I, Trounson A, Dawson G, Rogers P. Tripronuclear human oocytes: altered cleavage patterns and subsequent karyotypic analysis of embryos. Biol Reprod. 1987;37:395–401.
Meseguer M, Herrero J, Tejera A, Hilligsoe KM, Ramsing NB, Remohi J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26:2658–71.
Rubio I, Kuhlmann R, Agerholm I, Kirk J, Herrero J, Escriba MJ, et al. Limited implantation success of direct-cleaved human zygotes: a time-lapse study. Fertil Steril. 2012;98:1458–63.
Gleicher N, Vidali A, Braverman J, Kushnir VA, Barad DH, Hudson C, et al. Accuracy of preimplantation genetic screening (PGS) is compromised by degree of mosaicism of human embryos. Reprod Biol Endocrinol. 2016;14:54.
Liu J, Wang W, Sun X, Liu L, Jin H, Li M, et al. DNA microarray reveals that high proportions of human blastocysts from women of advanced maternal age are aneuploid and mosaic. Biol Reprod. 2012;87:148.
Baart EB, Martini E, Van Den Berg I, Macklon NS, Galjaard RJ, Fauser BC, et al. Preimplantation genetic screening reveals a high incidence of aneuploidy and mosaicism in embryos from young women undergoing IVF. Hum Reprod. 2006;21:223–33.
Scott RT Jr, Galliano D. The challenge of embryonic mosaicism in preimplantation genetic screening. Fertil Steril. 2016;105:1150–2.
Chow JF, Yeung WS, Lau EY, Lee VC, Ng EH, Ho PC. Array comparative genomic hybridization analyses of all blastomeres of a cohort of embryos from young IVF patients revealed significant contribution of mitotic errors to embryo mosaicism at the cleavage stage. Reprod Biol Endocrinol. 2014;12:105.
Wells D, Delhanty JD. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol Hum Reprod. 2000;6:1055–62.
Fragouli E, Alfarawati S, Spath K, Babariya D, Tarozzi N, Borini A, et al. Analysis of implantation and ongoing pregnancy rates following the transfer of mosaic diploid-aneuploid blastocysts. Hum Genet. 2017;136:805–19.
Palermo G, Munne S, Cohen J. The human zygote inherits its mitotic potential from the male gamete. Hum Reprod. 1994;9:1220–5.
Sathananthan AH, Tarin JJ, Gianaroli L, Ng SC, Dharmawardena V, Magli MC, et al. Development of the human dispermic embryo. Hum Reprod Update. 1999;5:553–60.
Gisselsson D, Jin Y, Lindgren D, Persson J, Gisselsson L, Hanks S, et al. Generation of trisomies in cancer cells by multipolar mitosis and incomplete cytokinesis. Proc Natl Acad Sci U S A. 2010;107:20489–93.
Chatzimeletiou K, Morrison EE, Prapas N, Prapas Y, Handyside AH. Spindle abnormalities in normally developing and arrested human preimplantation embryos in vitro identified by confocal laser scanning microscopy. Hum Reprod. 2005;20:672–82.
Acknowledgements
We thank Pro. William SB Yeung (The University of Hong Kong) for critical reading and editing of the manuscript.
Funding
This research was supported by the National Nature Science Foundation of China (81300549), Translational Medicine Program of Chinese PLA General Hospital (2016TM-028) and National Key Technology Support Program (2012BAI32B04).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The Institutional Review Board of Chinese PLA General Hospital (S2016-106-01) approved this study. All the recruited patients signed a written consent.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(XLSX 15 kb)
Rights and permissions
About this article
Cite this article
Ma, M., Zhang, S., Lu, C. et al. Chromosome constitution of equal-sized three-cell embryos using next-generation sequencing technology. J Assist Reprod Genet 36, 307–314 (2019). https://doi.org/10.1007/s10815-018-1362-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-018-1362-7