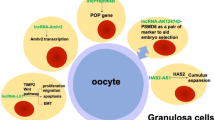
Abstract
Objective
To screen differentially expressed lncRNAs involved in OHSS. OHSS is defined as ovarian hyperstimulation syndrome. It is characterized as enlarged ovary and increased vascular permeability.
Design
Case-control study.
Setting
University-affiliated hospital.
Patient(s)
Patients with OHSS high risk (n = 30) and low risk (n = 30) were included in this study.
Intervention(s)
None.
Main outcome measure(s)
LncRNAs from women with OHSS high risk and low risk were used for high-throughput sequencing profiling. The eight most differentially expressed lncRNAs in granulosa cells were validated by semi-quantitative reverse transcription-polymerase chain reaction analysis.
Result(s)
A total of 23,815 lncRNAs were detected and 482 were differentially expressed (fold-change ≥2; p < 0.05, FDR value < 0.001), of which 205 were upregulated and 277 were downregulated. Lnc-SEC16B.1-6, lnc-SNURF-13, lnc-LGR6-6, and lnc-H2AFY2-2 were up-regulated, while lnc-BRD2-2, lnc-HSPA6-2, and lnc-CLIC6-5 were downregulated significantly in granulosa cells. These results were confirmed by qRT-PCR. KEGG pathways and Gene Ontology enrichment analysis revealed that several biological processes were significantly associated. Meanwhile, the lncRNA/miRNA interaction network was established according to ceRNA network model.
Conclusion(s)
Comprehensive expression screening identified eight novel lncRNAs associated with risk factors of OHSS process. Although it is unclear how these altered lncRNAs regulate the process of OHSS, our findings suggest these lncRNAs may be novel players in OHSS development.





Similar content being viewed by others
References
Luke B, Brown MB, Morbeck DE, Hudson SB, Coddington CC 3rd, Stern JE. Factors associated with ovarian hyperstimulation syndrome (OHSS) and its effect on assisted reproductive technology (ART) treatment and outcome. Fertil Steril. 2010;94:1399–404.
Mourad S, Brown J, Farquhar C. Interventions for the prevention of OHSS in ART cycles: an overview of Cochrane reviews. Cochrane Database Syst Rev. 2017;1:CD012103.
A.a.o. Practice Committee of the American Society for Reproductive Medicine. Electronic address, M. Practice Committee of the American Society for Reproductive. Prevention and treatment of moderate and severe ovarian hyperstimulation syndrome: a guideline. Fertil Steril. 2016;106:1634–47.
Humaidan P, Nelson SM, Devroey P, Coddington CC, Schwartz LB, Gordon K, et al. Ovarian hyperstimulation syndrome: review and new classification criteria for reporting in clinical trials. Hum Reprod. 2016;31:1997–2004.
Papanikolaou EG, Pozzobon C, Kolibianakis EM, Camus M, Tournaye H, Fatemi HM, et al. Incidence and prediction of ovarian hyperstimulation syndrome in women undergoing gonadotropin-releasing hormone antagonist in vitro fertilization cycles. Fertil Steril. 2006;85:112–20.
Delvigne A, Rozenberg S. Epidemiology and prevention of ovarian hyperstimulation syndrome (OHSS): a review. Hum Reprod Update. 2002;8:559–77.
Cerrillo M, Pacheco A, Rodriguez S, Gomez R, Delgado F, Pellicer A, et al. Effect of GnRH agonist and hCG treatment on VEGF, angiopoietin-2, and VE-cadherin: trying to explain the link to ovarian hyperstimulation syndrome. Fertil Steril. 2011;95:2517–9.
Naredi N, Talwar P, Sandeep K. VEGF antagonist for the prevention of ovarian hyperstimulation syndrome: current status. Med J Armed Forces India. 2014;70:58–63.
Pietrowski D, Szabo L, Sator M, Just A, Egarter C. Ovarian hyperstimulation syndrome is correlated with a reduction of soluble VEGF receptor protein level and a higher amount of VEGF-A. Hum Reprod. 2012;27:196–9.
Scotti L, Abramovich D, Pascuali N, Irusta G, Meresman G, Tesone M, et al. Local VEGF inhibition prevents ovarian alterations associated with ovarian hyperstimulation syndrome. J Steroid Biochem Mol Biol. 2014;144(Pt B):392–401.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14.
Xu B, Zhang YW, Tong XH, Liu YS. Characterization of microRNA profile in human cumulus granulosa cells: identification of microRNAs that regulate notch signaling and are associated with PCOS. Mol Cell Endocrinol. 2015;404:26–36.
Jiang L, Huang J, Li L, Chen Y, Chen X, Zhao X, et al. MicroRNA-93 promotes ovarian granulosa cells proliferation through targeting CDKN1A in polycystic ovarian syndrome. J Clin Endocrinol Metab. 2015;100:E729–38.
Wang H, Cao Q, Ge J, Liu C, Ma Y, Meng Y, et al. LncRNA-regulated infection and inflammation pathways associated with pregnancy loss: genome wide differential expression of lncRNAs in early spontaneous abortion. Am J Reprod Immunol. 2014;72:359–75.
Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. elife. 2013;2:e00762.
Moran I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16:435–48.
Qiu JJ, Ye LC, Ding JX, Feng WW, Jin HY, Zhang Y, et al. Expression and clinical significance of estrogen-regulated long non-coding RNAs in estrogen receptor alpha-positive ovarian cancer progression. Oncol Rep. 2014;31:1613–22.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36.
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5.
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–22.
Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–53.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–83.
Shen L, Wang Q, Liu R, Chen Z, Zhang X, Zhou P, et al. LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res. 2018;46(2):717–29.
Scotti L, Irusta G, Abramovich D, Tesone M, Parborell F. Administration of a gonadotropin-releasing hormone agonist affects corpus luteum vascular stability and development and induces luteal apoptosis in a rat model of ovarian hyperstimulation syndrome. Mol Cell Endocrinol. 2011;335:116–25.
Chen CD, Chen HF, Lu HF, Chen SU, Ho HN, Yang YS. Value of serum and follicular fluid cytokine profile in the prediction of moderate to severe ovarian hyperstimulation syndrome. Hum Reprod. 2000;15:1037–42.
Bedarida GV, Hoffmann U, Tato F. Jugular vein thrombosis with severe local and systemic inflammation in a woman with ovarian hyperstimulation syndrome. Thromb Haemost. 2006;95:1035–7.
Gera PS, Tatpati LL, Allemand MC, Wentworth MA, Coddington CC. Ovarian hyperstimulation syndrome: steps to maximize success and minimize effect for assisted reproductive outcome. Fertil Steril. 2010;94:173–8.
Yan Z, Shah PK, Amin SB, Samur MK, Huang N, Wang X, et al. Integrative analysis of gene and miRNA expression profiles with transcription factor-miRNA feed-forward loops identifies regulators in human cancers. Nucleic Acids Res. 2012;40:e135.
Murri M, Insenser M, Fernandez-Duran E, San-Millan JL, Escobar-Morreale HF. Effects of polycystic ovary syndrome (PCOS), sex hormones, and obesity on circulating miRNA-21, miRNA-27b, miRNA-103, and miRNA-155 expression. J Clin Endocrinol Metab. 2013;98:E1835–44.
Huang X, Liu C, Hao C, Tang Q, Liu R, Lin S, et al. Identification of altered microRNAs and mRNAs in the cumulus cells of PCOS patients: miRNA-509-3p promotes oestradiol secretion by targeting MAP3K8. Reproduction. 2016;151:643–55.
Acknowledgements
This study was funded by National Natural Science Funding (no. 81701519).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(XLS 169 kb)
Rights and permissions
About this article
Cite this article
Lin, H., Li, Y., Xing, W. et al. Genome-wide screening differential long non-coding RNAs expression profiles discloses its roles involved in OHSS development. J Assist Reprod Genet 35, 1473–1482 (2018). https://doi.org/10.1007/s10815-018-1199-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-018-1199-0