Abstract
Purpose
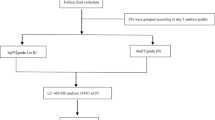
The etiology of maternal aging, a common cause of female factor infertility and a rate-limiting step in vitro fertilization (IVF) success, remains still unclear. Proteomic changes responsible for the impaired successful pregnancy outcome after IVF with aged blastocysts have not been yet evaluated. The objective of this prospective study was to employ proteomic techniques and bioinformatic tools to enlight differences at the protein level in blastocoel fluid of aged and younger woman.
Methods
Protein composition of human blastocoel fluid isolated by micromanipulation from 46 blastocysts of women aged <37 years (group A) and 29 of women aged ≥37 years (group B) have been identified by a shotgun proteomic approach based on high-resolution nano-liquid chromatography electrospray-ionization-tandem mass spectrometry (nLC-ESI-MS/MS) using label free for the relative quantification of their expression levels.
Results
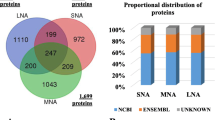
The proteomic analysis leads to the identification and quantification of 148 proteins; 132 and 116 proteins were identified in groups A and B, respectively. Interestingly, the identified proteins are mainly involved in processes aimed at fine tuning embryo implantation and development. Among the 100 proteins commonly expressed in both groups, 17 proteins are upregulated and 44 downregulated in group B compared to group A. Overall, the analysis identified 33 proteins, which were increased or present only in B while 76 were decreased in B or present only in A.
Conclusions
Data revealed that maternal aging mainly affects blastocyst survival and implantation through unbalancing the equilibrium of the ubiquitin system known to play a crucial role in fine-tuning several aspects required to ensure successful pregnancy outcome.




Similar content being viewed by others
References
Paiva P, Salamonsen LA, Manuelpillai U, Dimitriadis E. Interleukin 11 inhibits human trophoblast invasion indicating a likely role in the decidual restraint of trophoblast invasion during placentation. Biol Reprod. 2009;80(2):302–10. doi:10.1095/biolreprod.108.071415.
Koot YE, Teklenburg G, Salker MS, Brosens JJ, Macklon NS. Molecular aspects of implantation failure. Biochim Biophys Acta. 2012;1822(12):1943–50. doi:10.1016/j.bbadis.2012.05.017.
Dimitriadis E, White CA, Jones RL, Salamonsen LA. Cytokines, chemokines and growth factors in endometrium related to implantation. Hum Reprod Update. 2005;11(6):613–30. doi:10.1093/humupd/dmi023.
Humm KC, Dodge LE, Wu LH, Penzias AS, Malizia BA, Sakkas D, et al. In vitro fertilization in women under 35: counseling should differ by age. J Assist Reprod Genet. 2015. doi:10.1007/s10815-015-0570-7.
Malizia BA, Hacker MR, Penzias AS. Cumulative live-birth rates after in vitro fertilization. N Engl J Med. 2009;360(3):236–43. doi:10.1056/NEJMoa0803072.
Luke B, Brown MB, Wantman E, Lederman A, Gibbons W, Schattman GL, et al. Cumulative birth rates with linked assisted reproductive technology cycles. N Engl J Med. 2012;366(26):2483–91. doi:10.1056/NEJMoa1110238.
Aplin JD. Embryo implantation: the molecular mechanism remains elusive. Reprod Biomed Online. 2006;13(6):833–9.
Garrido-Gomez T, Dominguez F, Simon C. Proteomics of embryonic implantation. Handb Exp Pharmacol. 2010;198:67–78. doi:10.1007/978-3-642-02062-9_5.
Aydiner F, Yetkin CE, Seli E. Perspectives on emerging biomarkers for non-invasive assessment of embryo viability in assisted reproduction. Curr Mol Med. 2010;10(2):206–15.
Bromer JG, Seli E. Assessment of embryo viability in assisted reproductive technology: shortcomings of current approaches and the emerging role of metabolomics. Curr Opin Obstetr Gynecol. 2008;20(3):234–41. doi:10.1097/GCO.0b013e3282fe723d.
Latham KE, Garrels JI, Chang C, Solter D. Analysis of embryonic mouse development: construction of a high-resolution, two-dimensional gel protein database. Appl Theoret Electrophoresis : Off J Int Electrophoresis Soc. 1992;2(6):163–70.
Shi CZ, Collins HW, Garside WT, Buettger CW, Matschinsky FM, Heyner S. Protein databases for compacted eight-cell and blastocyst-stage mouse embryos. Mol Reprod Dev. 1994;37(1):34–47. doi:10.1002/mrd.1080370106.
Katz-Jaffe MG, Linck DW, Schoolcraft WB, Gardner DK. A proteomic analysis of mammalian preimplantation embryonic development. Reproduction. 2005;130(6):899–905. doi:10.1530/rep.1.00854.
Katz-Jaffe MG, Gardner DK, Schoolcraft WB. Proteomic analysis of individual human embryos to identify novel biomarkers of development and viability. Fertil Steril. 2006;85(1):101–7. doi:10.1016/j.fertnstert.2005.09.011.
Katz-Jaffe MG, Schoolcraft WB, Gardner DK. Analysis of protein expression (secretome) by human and mouse preimplantation embryos. Fertil Steril. 2006;86(3):678–85. doi:10.1016/j.fertnstert.2006.05.022.
Poli M, Ori A, Child T, Jaroudi S, Spath K, Beck M et al. Characterization and quantification of proteins secreted by single human embryos prior to implantation. EMBO Molec Med. 2015; 7(11): 1465–79. doi: 10.15252/emmm.201505344.
Jensen PL, Beck HC, Petersen J, Hreinsson J, Wanggren K, Laursen SB, et al. Proteomic analysis of human blastocoel fluid and blastocyst cells. Stem Cells Dev. 2013;22(7):1126–35. doi:10.1089/scd.2012.0239.
Katz-Jaffe MG, McReynolds S, Gardner DK, Schoolcraft WB. The role of proteomics in defining the human embryonic secretome. Mol Hum Reprod. 2009;15(5):271–7. doi:10.1093/molehr/gap012.
Gardner RL, Beddington RS. Multi-lineage ‘stem’ cells in the mammalian embryo. J Cell Sci Suppl. 1988;10:11–27.
Chambery A, Colucci-D’Amato L, Vissers JP, Scarpella S, Langridge JI, Parente A. Proteomic profiling of proliferating and differentiated neural mes-c-myc A1 cell line from mouse embryonic mesencephalon by LC-MS. J Proteome Res. 2009;8(1):227–38. doi:10.1021/pr800454n.
Levin Y, Wang L, Ingudomnukul E, Schwarz E, Baron-Cohen S, Palotas A, et al. Real-time evaluation of experimental variation in large-scale LC-MS/MS-based quantitative proteomics of complex samples. J Chromatogr B Anal Technol Biomed Life Sci. 2009;877(13):1299–305. doi:10.1016/j.jchromb.2008.11.007.
Levi-Setti PE, Menduni F, Smeraldi A, Patrizio P, Morenghi E, Albani E. Artificial shrinkage of blastocysts prior to vitrification improves pregnancy outcome: analysis of 1028 consecutive warming cycles. J Assist Reprod Genet. 2016;33(4):461–6. doi:10.1007/s10815-016-0655-y.
Vernocchi V, Morselli MG, Varesi S, Nonnis S, Maffioli E, Negri A, et al. Sperm ubiquitination in epididymal feline semen. Theriogenology. 2014;82(4):636–42. doi:10.1016/j.theriogenology.2014.06.002.
Maffioli EAF, Nonnis S, Santagata F, Negri A, DeLano FA, Santamaria MH, Kistler EB, Schmid-Schönbein GW, Tedeschi G. Analysis of rat plasma peptidome in hemorrhagic shock. Shock. 2015; 44 (Suppl 2:6).
Tamplenizza M, Lenardi C, Maffioli E, Nonnis S, Negri A, Forti S, et al. Nitric oxide synthase mediates PC12 differentiation induced by the surface topography of nanostructured TiO2. J Nanobiotechnol. 2013;11:35. doi:10.1186/1477-3155-11-35.
Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–72. doi:10.1038/nbt.1511.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–52. doi:10.1093/nar/gku1003.
Fraser KB, Moehle MS, Daher JP, Webber PJ, Williams JY, Stewart CA, et al. LRRK2 secretion in exosomes is regulated by 14-3-3. Hum Mol Genet. 2013;22(24):4988–5000. doi:10.1093/hmg/ddt346.
Vassena R, Boue S, Gonzalez-Roca E, Aran B, Auer H, Veiga A, et al. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development. 2011;138(17):3699–709. doi:10.1242/dev.064741.
Bebington C, Doherty FJ, Fleming SD. The possible biological and reproductive functions of ubiquitin. Hum Reprod Update. 2001;7(1):102–11.
Bebington C, Bell SC, Doherty FJ, Fazleabas AT, Fleming SD. Localization of ubiquitin and ubiquitin cross-reactive protein in human and baboon endometrium and decidua during the menstrual cycle and early pregnancy. Biol Reprod. 1999;60(4):920–8.
Bebington C, Doherty FJ, Fleming SD. Ubiquitin cross-reactive protein gene expression is increased in decidualized endometrial stromal cells at the initiation of pregnancy. Mol Hum Reprod. 1999;5(10):966–72.
Bebington C, Doherty FJ, Ndukwe G, Fleming SD. The progesterone receptor and ubiquitin are differentially regulated within the endometrial glands of the natural and stimulated cycle. Mol Hum Reprod. 2000;6(3):264–8.
Grzmil P, Altmann ME, Adham IM, Engel U, Jarry H, Schweyer S, et al. Embryo implantation failure and other reproductive defects in Ube2q1-deficient female mice. Reproduction. 2013;145(1):45–56. doi:10.1530/REP-12-0054.
Sutovsky P, Motlik J, Neuber E, Pavlok A, Schatten G, Palecek J, et al. Accumulation of the proteolytic marker peptide ubiquitin in the trophoblast of mammalian blastocysts. Cloning Stem Cells. 2001;3(3):157–61. doi:10.1089/153623001753205115.
Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci U S A. 2003;100(5):2468–73. doi:10.1073/pnas.0434312100.
Leeb M, Wutz A. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol. 2007;178(2):219–29. doi:10.1083/jcb.200612127.
Fenner BJ, Scannell M, Prehn JH. Identification of polyubiquitin binding proteins involved in NF-kappaB signaling using protein arrays. Biochim Biophys Acta. 2009;1794(7):1010–6. doi:10.1016/j.bbapap.2009.02.013.
Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9(6):769–79. doi:10.1016/j.devcel.2005.10.007.
Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435(7042):687–92. doi:10.1038/nature03588.
de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7(5):663–76. doi:10.1016/j.devcel.2004.10.005.
Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431(7010):873–8. doi:10.1038/nature02985.
Endoh M, Endo TA, Endoh T, Isono K, Sharif J, Ohara O, et al. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012;8(7):e1002774. doi:10.1371/journal.pgen.1002774.
Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol. 2014;21(6):569–71. doi:10.1038/nsmb.2833.
Luo M, Zhou J, Leu NA, Abreu CM, Wang J, Anguera MC, et al. Polycomb protein SCML2 associates with USP7 and counteracts histone H2A ubiquitination in the XY chromatin during male meiosis. PLoS Genet. 2015;11(1):e1004954. doi:10.1371/journal.pgen.1004954.
Qin J, Whyte WA, Anderssen E, Apostolou E, Chen HH, Akbarian S, et al. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell. 2012;11(3):319–32. doi:10.1016/j.stem.2012.06.002.
Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–53. doi:10.1038/nature04733.
Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(7286):306–10. doi:10.1038/nature08788.
O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21(13):4330–6. doi:10.1128/MCB.21.13.4330-4336.2001.
Kerppola TK. Polycomb group complexes—many combinations, many functions. Trends Cell Biol. 2009;19(12):692–704. doi:10.1016/j.tcb.2009.10.001.
Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. doi:10.1038/nrm2763.
Gong Y, Yue J, Wu X, Wang X, Wen J, Lu L, et al. NSPc1 is a cell growth regulator that acts as a transcriptional repressor of p21Waf1/Cip1 via the RARE element. Nucleic Acids Res. 2006;34(21):6158–69. doi:10.1093/nar/gkl834.
Cortezzi SS, Garcia JS, Ferreira CR, Braga DP, Figueira RC, Iaconelli Jr A, et al. Secretome of the preimplantation human embryo by bottom-up label-free proteomics. Anal Bioanal Chem. 2011;401(4):1331–9. doi:10.1007/s00216-011-5202-1.
Nashun B, Yukawa M, Liu H, Akiyama T, Aoki F. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development. 2010;137(22):3785–94. doi:10.1242/dev.051805.
Wu BJ, Dong FL, Ma XS, Wang XG, Lin F, Liu HL. Localization and expression of histone H2A variants during mouse oogenesis and preimplantation embryo development. Genet Molec Res : GMR. 2014;13(3):5929–39. doi:10.4238/2014.August.7.8.
Singh RK, Liang D, Gajjalaiahvari UR, Kabbaj MH, Paik J, Gunjan A. Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell Cycle. 2010;9(20):4236–44.
Liu Y, Tseng M, Perdreau SA, Rossi F, Antonescu C, Besmer P, et al. Histone H2AX is a mediator of gastrointestinal stromal tumor cell apoptosis following treatment with imatinib mesylate. Cancer Res. 2007;67(6):2685–92. doi:10.1158/0008-5472.CAN-06-3497.
Liu Y, Parry JA, Chin A, Duensing S, Duensing A. Soluble histone H2AX is induced by DNA replication stress and sensitizes cells to undergo apoptosis. Mol Cancer. 2008;7:61. doi:10.1186/1476-4598-7-61.
Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11(8):925–33. doi:10.1038/ncb1903.
Singh RK, Paik J, Gunjan A. Generation and management of excess histones during the cell cycle. Front Biosci (Landmark Ed). 2009;14:3145–58.
Cook AJ, Gurard-Levin ZA, Vassias I, Almouzni G. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol Cell. 2011;44(6):918–27. doi:10.1016/j.molcel.2011.11.021.
Li Y, Lu J, Prochownik EV. Dual role for SUMO E2 conjugase Ubc9 in modulating the transforming and growth-promoting properties of the HMGA1b architectural transcription factor. J Biol Chem. 2007;282(18):13363–71. doi:10.1074/jbc.M610919200.
Benbrook DM, Long A. Integration of autophagy, proteasomal degradation, unfolded protein response and apoptosis. Exp Oncol. 2012;34(3):286–97.
Murray P, Edgar D. Regulation of programmed cell death by basement membranes in embryonic development. J Cell Biol. 2000;150(5):1215–21.
Carmignac V, Svensson M, Korner Z, Elowsson L, Matsumura C, Gawlik KI, et al. Autophagy is increased in laminin alpha2 chain-deficient muscle and its inhibition improves muscle morphology in a mouse model of MDC1A. Hum Mol Genet. 2011;20(24):4891–902. doi:10.1093/hmg/ddr427.
Taylor AH, Amoako AA, Bambang K, Karasu T, Gebeh A, Lam PM, et al. Endocannabinoids and pregnancy. Clin Chimica Acta; Int J Clin Chem. 2010;411(13-14):921–30. doi:10.1016/j.cca.2010.03.012.
Maccarrone M. Endocannabinoids: friends and foes of reproduction. Prog Lipid Res. 2009;48(6):344–54. doi:10.1016/j.plipres.2009.07.001.
Battista N, Bari M, Maccarrone M. Endocannabinoids and reproductive events in health and disease. Handb Exp Pharmacol. 2015;231:341–65. doi:10.1007/978-3-319-20825-1_12.
McKinney MK, Cravatt BF. Structure and function of fatty acid amide hydrolase. Annu Rev Biochem. 2005;74:411–32. doi:10.1146/annurev.biochem.74.082803.133450.
Rapino C, Battista N, Bari M, Maccarrone M. Endocannabinoids as biomarkers of human reproduction. Hum Reprod Update. 2014;20(4):501–16. doi:10.1093/humupd/dmu004.
Maccarrone M, Bisogno T, Valensise H, Lazzarin N, Fezza F, Manna C, et al. Low fatty acid amide hydrolase and high anandamide levels are associated with failure to achieve an ongoing pregnancy after IVF and embryo transfer. Mol Hum Reprod. 2002;8(2):188–95.
Maccarrone M, Valensise H, Bari M, Lazzarin N, Romanini C, Finazzi-Agro A. Relation between decreased anandamide hydrolase concentrations in human lymphocytes and miscarriage. Lancet. 2000;355(9212):1326–9. doi:10.1016/S0140-6736(00)02115-2.
Habayeb OM, Taylor AH, Finney M, Evans MD, Konje JC. Plasma anandamide concentration and pregnancy outcome in women with threatened miscarriage. Jama. 2008;299(10):1135–6. doi:10.1001/jama.299.10.1135.
Wei BQ, Mikkelsen TS, McKinney MK, Lander ES, Cravatt BF. A second fatty acid amide hydrolase with variable distribution among placental mammals. J Biol Chem. 2006;281(48):36569–78. doi:10.1074/jbc.M606646200.
Jiang JY, Xiong H, Cao M, Xia X, Sirard MA, Tsang BK. Mural granulosa cell gene expression associated with oocyte developmental competence. J Ovarian Res. 2010;3:6. doi:10.1186/1757-2215-3-6.
Esposito G, Schiattarella GG, Perrino C, Cattaneo F, Pironti G, Franzone A, et al. Dermcidin: a skeletal muscle myokine modulating cardiomyocyte survival and infarct size after coronary artery ligation. Cardiovasc Res. 2015;107(4):431–41. doi:10.1093/cvr/cvv173.
Huang B, Porter G. Expression of proline-rich Akt-substrate PRAS40 in cell survival pathway and carcinogenesis. Acta Pharmacol Sin. 2005;26(10):1253–8. doi:10.1111/j.1745-7254.2005.00184.x.
O’Neill C, Li Y, Jin XL. Survival signaling in the preimplantation embryo. Theriogenology. 2012;77(4):773–84. doi:10.1016/j.theriogenology.2011.12.016.
Abbas W, Kumar A, Herbein G. The eEF1A proteins: at the crossroads of oncogenesis, apoptosis, and viral infections. Front Oncol. 2015;5:75. doi:10.3389/fonc.2015.00075.
Migliaccio N, Ruggiero I, Martucci NM, Sanges C, Arbucci S, Tate R, et al. New insights on the interaction between the isoforms 1 and 2 of human translation elongation factor 1A. Biochimie. 2015. doi:10.1016/j.biochi.2015.07.021.
Acknowledgements
We thank Dr. Francesca Grassi Scalvini and Dr. Fabiana Santagata for their skillful technical assistance.
Authors’ roles
G.T., E.A., V.P., A.B., and P.E.L.S. designed the research. G.T., E.A., V.P., A.B., E.M., A.N., and S.N. performed the research. G.T., E.A., V.P., A.B., E.M.B., E.M., A.N., S.N., and M.M. analyzed the data. G.T., E.A., V.P., A.B., E.M.B., and M.M. wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
None declared.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Capsule
Maternal aging mainly affects blastocyst survival and implantation through unbalancing the equilibrium of theubiquitin system
Gabriella Tedeschi and Elena Albani contributed equally to this work.
Rights and permissions
About this article
Cite this article
Tedeschi, G., Albani, E., Borroni, E.M. et al. Proteomic profile of maternal-aged blastocoel fluid suggests a novel role for ubiquitin system in blastocyst quality. J Assist Reprod Genet 34, 225–238 (2017). https://doi.org/10.1007/s10815-016-0842-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-016-0842-x