Abstract
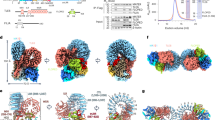
The subcortical maternal complex (SCMC) is a multiprotein complex uniquely expressed in mammalian oocytes and early embryos, essential for zygote progression beyond the first embryonic cell divisions. Similiar to other factors encoded by maternal effect genes, the physiological role of SCMC remains unclear, although recent evidence has provided important molecular insights into different possible functions. Its potential involvement in human fertility is attracting increasing attention; however, the complete story is far from being told. The present mini review provides an overview of recent findings related to the SCMC and discusses its potential physiological role/s with the aim of inspiring new directions for future research.


Similar content being viewed by others
References
Inhorn MC, Patrizio P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum Reprod Update. 2015;21(4):411–26.
Tarín JJ, García-Pérez MA, Cano A. Assisted reproductive technology results: why are live-birth percentages so low? Mol Reprod Dev. 2014;81:568–83.
Alazami AM, Awad SM, Coskun S, Al-Hassan S, Hijazi H, Abdulwahab FM, et al. TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 2015;16:240.
Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, et al. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet. 2000;26:267–8.
Wu X, Viveiros MM, Eppig JJ, Bai Y, Fitzpatrick SL, Matzuk MM. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet. 2003;33:187–91.
Dean J. Oocyte-specific genes regulate follicle formation, fertility and early mouse development. J Reprod Immunol. 2002;53:171–80.
Zheng P, Dean J. Oocyte-specific genes affect folliculogenesis, fertilization, and early development. Semin Reprod Med. 2007;25:243–51.
Zhang K, Smith GW. Maternal control of early embryogenesis in mammals. Reprod Fertil Dev. 2015;27:880–96.
Yu C, Ji SY, Sha QQ, Dang Y, Zhou JJ, Zhang YL, et al. BTG4 is a meiotic cell cycle-coupled maternal-zygotic-transition licensing factor in oocytes. Nat Struct Mol Biol. 2016;23:387–94.
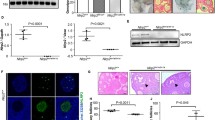
Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15:416–25.
Yu XJ, Yi Z, Gao Z, Qin D, Zhai Y, Chen X, et al. The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics. Nat Commun. 2014;5:4887.
Zhu K, Yan L, Zhang X, Lu X, Wang T, Yan J, et al. Identification of a human subcortical maternal complex. Mol Hum Reprod. 2015;21:320–9.
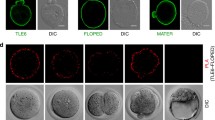
Ohsugi M, Zheng P, Baibakov B, Li L, Dean J. Maternally derived FILIAMATER complex localizes asymmetrically in cleavage-stage mouse embryos. Development. 2008;135:259–69.
Tashiro F, Kanai-Azuma M, Miyazaki S, Kato M, Tanaka T, Toyoda S, et al. Maternal-effect gene Ces5/Ooep/Moep19/Floped is essential for oocyte cytoplasmic lattice formation and embryonic development at the maternal-zygotic stage transition. Genes Cells. 2010;15:813–28.
Zheng P, Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc Natl Acad Sci U S A. 2009;106:7473–8.
Herr JC, Chertihin O, Digilio L, Jha KN, Vemuganti S, Flickinger CJ. Distribution of RNA binding protein MOEP19 in the oocyte cortex and early embryo indicates pre-patterning related to blastomere polarity and trophectoderm specification. Dev Biol. 2008;314:300–16.
Tong ZB, Gold L, De Pol A, Vanevski K, Dorward H, Sena P, et al. Developmental expression and subcellular localization of mouse MATER, an oocyte specific-protein essential for early development. Endocrinology. 2004;145:1427–34.
Tong ZB, Nelson LM. A mouse gene encoding an oocyte antigen associated with autoimmune premature ovarian failure. Endocrinology. 1999;140:3720–6.
Bebbere D, Ariu F, Bogliolo L, Masala L, Murrone O, Fattorini M, et al. Expression of maternally derived KHDC3, NLRP5, OOEP and TLE6 is associated with oocyte developmental competence in the ovine species. BMC Dev Biol. 2014;14:40.
Adona PR, de Bem TH, Mesquita LG, Rochetti RC, Leal CL. Embryonic development and gene expression in oocytes cultured in vitro in supplemented pre-maturation and maturation media. Reprod Domest Anim. 2011;46:e31–8.
Romar R, De Santis T, Papillier P, Perreau C, Thélie A, Dell’Aquila ME, et al. Expression of maternal transcripts during bovine oocyte in vitro maturation is affected by donor age. Reprod Domest Anim. 2011;46:e23–30.
Pisani LF, Ramelli P, Lazzari B, Braglia S, Ceciliani F, Mariani P. Characterization of maternal antigen that embryos require (MATER/NLRP5) gene and protein in pig somatic tissues and germ cells. J Reprod Dev. 2010;56(1):41–8.
Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010;20:187–95.
Zheng P, Baibakov B, Wang XH, Dean J. PtdIns(3,4,5)P3 is constitutively synthesized and required for spindle translocation during meiosis in mouse oocytes. J Cell Sci. 2013;126:715–21.
Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius KJ, Jarvius J, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000.
Wright PW, Bolling LC, Calvert ME, Sarmento OF, Berkeley EV, Shea MC, et al. ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev Biol. 2003;256:73–88.
Esposito G, Vitale AM, Leijten FP, Strik AM, Koonen-Reemst AM, Yurttas P, et al. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol. 2007;273:25–31.
Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, et al. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135:2627–36.
Capco DG, Gallicano GI, McGaughey RW, Downing KH, Larabell CA. Cytoskeletal sheets of mammalian eggs and embryos: a lattice-like network of intermediate filaments. Cell Motil Cytoskeleton. 1993;24:85–99.
Wassarman PM, Josefowicz WJ. Oocyte development in the mouse: an ultrastructural comparison of oocytes isolated at various stages of growth and meiotic competence. J Morphol. 1978;156:209–35.
Gallicano GI, McGaughey RW, Capco DG. Cytoskeleton of the mouse egg and embryo: reorganization of planar elements. Cell Motil Cytoskeleton. 1991;18:143–54.
Kim B, Kan R, Anguish L, Nelson LM, Coonrod SA. Potential role for MATER in cytoplasmic lattice formation in murine oocytes. PLoS ONE. 2010;5(9), e12587.
Reid DW, Nicchitta CV. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2015;16:221–31.
Kang MK, Han SJ. Post-transcriptional and post-translational regulation during mouse oocyte maturation. BMB Rep. 2011;44:147–57.
Clarke HJ. Post-transcriptional control of gene expression during mouse oogenesis. Results Probl Cell Differ. 2012;55:1–21.
Weill L, Belloc E, Bava FA, Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol. 2012;19:577–85.
Sternlicht AL, Schultz RM. Biochemical studies of mammalian oogenesis: kinetics of accumulation of total and poly(A)-containing RNA during growth of the mouse oocyte. J Exp Zool. 1981;215:191–200.
Pierre A, Gautier M, Callebaut I, Bontoux M, Jeanpierre E, Pontarotti P, et al. Atypical structure and phylogenomic evolution of the new eutherian oocyte- and embryo-expressed KHDC1/DPPA5/ECAT1/OOEP gene family. Genomics. 2007;90:583–94.
Wang J, Xu M, Zhu K, Li L, Liu X. The N-terminus of FILIA forms an atypical KH domain with a unique extension involved in interaction with RNA. PLoS ONE. 2012;7, e30209.
Susor A, Jansova D, Cerna R, Danylevska A, Anger M, Toralova T, et al. Temporal and spatial regulation of translation in the mammalian oocyte via the mTOR-eIF4F pathway. Nat Commun. 2015;6:6078.
Joshi S, Davies H, Sims LP, Levy SE, Dean J. Ovarian gene expression in the absence of FIGLA, an oocyte-specific transcription factor. BMC Dev Biol. 2007;7:67.
Park MW, Kim KH, Kim EY, Lee SY, Ko JJ, Lee KA. Associations among Sebox and other MEGs and its effects on early embryogenesis. PLoS ONE. 2015;10(2), e0115050.
Liang L, Soyal SM, Dean J. FIGalpha, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development. 1997;124:4939–47.
Fernandes R, Tsuda C, Perumalsamy AL, Naranian T, Chong J, Acton BM, et al. NLRP5 mediates mitochondrial function in mouse oocytes and embryos. Biol Reprod. 2012;86:138,1–10.
Dumollard R, Duchen M, Carroll J. The role of mitochondrial function in the oocyte and embryo. Curr Top Dev Biol. 2007;77:21–49.
Muggleton-Harris AL, Brown JJ. Cytoplasmic factors influence mitochondrial reorganization and resumption of cleavage during culture of early mouse embryos. Hum Reprod. 1988;3:1020–8.
Kabashima K, Matsuzaki M, Suzuki H. Both microtubules and microfilaments mutually control the distribution of mitochondria in two cell embryos of golden hamsters. J Mamm Ova Res. 2007;24:120–5.
Docherty LE, Rezwan FI, Poole RL, Turner CL, Kivuva E, Maher ER, et al. Mutations in NLRP5 are associated with reproductive wastage and multilocus imprinting disorders in humans. Nat Commun. 2015;6:8086.
Akoury E, Zhang L, Ao A, Slim R. NLRP7andKHDC3L, the two maternal effect proteins responsible for recurrent hydatidiform moles, co-localize to the oocyte cytoskeleton. Hum Reprod. 2015;30:159–69.
Hanna CW, Kelsey G. The specification of imprints in mammals. Heredity. 2014;113:176–83.
Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet. 2014;15:517–30.
Arnaud P, Feil R. Epigenetic deregulation of genomic imprinting in human 21 disorders and following assisted reproduction. Birth Defects Res C Embryo Today. 2005;75:81–97.
Scarano MI, Strazzullo M, Matarazzo MR, D’Esposito M. DNA methylation 40 years later: its role in human health and disease. J Cell Physiol. 2005;204:21–35.
Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, Kuick R, et al. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet. 2006;38:300–2.
El-Maarri O, Seoud M, Coullin P, Herbiniaux U, Oldenburg J, Rouleau G, et al. Maternal alleles acquiring paternal methylation patterns in biparental complete hydatidiform moles. Hum Mol Genet. 2003;12:1405–13.
Judson H, Hayward BE, Sheridan E, Bonthron DT. A global disorder of imprinting in the human female germ line. Nature. 2002;416:539–42.
Parry DA, Logan CV, Hayward BE, Shires M, Landolsi H, Diggle C, et al. Mutations causing familial biparental hydatidiform mole implicate c6orf221 as a possible regulator of genomic imprinting in the human oocyte. Am J Hum Genet. 2011;89:451–8.
Fallahian M, Sebire NJ, Savage PM, Seckl MJ, Fisher RA. Mutations in NLRP7 and KHDC3L confer a complete hydatidiform mole phenotype on digynic triploid conceptions. Hum Mutat. 2013;34:301–8.
Reddy R, Akoury E, Phuong Nguyen NM, Abdul-Rahman OA, Dery C, Gupta N, et al. Report of four new patients with protein-truncating mutations in C6orf221/ KHDC3L and colocalization with NLRP7. Eur J Hum Genet. 2013;21:957–64.
Qian J, Cheng Q, Murdoch S, Xu C, Jin F, Chebaro W, et al. The genetics of recurrent hydatidiform moles in China: correlations between NLRP7 mutations, molar genotypes and reproductive outcomes. Mol Hum Reprod. 2011;17:612–9.
Estrada H, Buentello B, Zenteno JC, Fiszman R, Aguinaga M. The p.L750V mutation in the NLRP7 gene is frequent in Mexican patients with recurrent molar pregnancies and is not associated with recurrent pregnancy loss. Prenat Diagn. 2013;33:205–8.
Kou YC, Shao L, Peng HH, Rosetta R, del Gaudio D, Wagner AF, et al. A recurrent intragenic genomic duplication, other novel mutations in NLRP7 and imprinting defects in recurrent biparental hydatidiform moles. Mol Hum Reprod. 2008;14:33–40.
Hayward BE, De Vos M, Talati N, Abdollahi MR, Taylor GR, Meyer E, et al. Genetic and epigenetic analysis of recurrent hydatidiform mole. Hum Mutat. 2009;30:E629–39.
Urrego R, Herrera-Puerta E, Chavarria NA, Camargo O, Wrenzycki C, Rodriguez-Osorio N. Follicular progesterone concentrations and messenger RNA expression of MATER and OCT-4 in immature bovine oocytes as predictors of developmental competence. Theriogenology. 2015;83:1179–87.
Trapphoff T, Heiligentag M, Dankert D, Demond H, Deutsch D, Fröhlich T, Arnold GJ, Grümmer R, Horsthemke B, Eichenlaub-Ritter U. Postovulatory aging affects dynamics of mRNA, expression and localization of maternal effect proteins, spindle integrity and pericentromeric proteins in mouse oocytes. Hum Reprod. 2016;31(1):133–49
Jiao ZX, Woodruff TK. Follicle microenvironment-associated alterations in gene expression in the mouse oocyte and its polar body. Fertil Steril. 2013;99:1453–9.
Dankert D, Demond H, Trapphoff T, Heiligentag M, Rademacher K, Eichenlaub-Ritter U, et al. Pre- and postovulatory aging of murine oocytes affect the transcript level and Poly(A) tail length of maternal effect genes. PLoS ONE. 2014;9(10), e108907.
Lu Y, He X, Zheng P. Decrease in expression of maternal effect gene Mater is associated with maternal ageing in mice. Mol. Hum. Reprod. 2016;0; 1–9.
Sharov AA, Falco G, Piao Y, Poosala S, Becker KG, Zonderman AB, et al. Effects of aging and calorie restriction on the global gene expression profiles of mouse testis and ovary. BMC Biol. 2008;6:24.
Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, Sharov AA, et al. Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Genet. 2004;13:2263–78.
Zhang P, Dixon M, Zucchelli M, Hambiliki F, Levkov L, Hovatta O, et al. Expression analysis of the NLRP gene family suggests a role in human preimplantation development. PLoS ONE. 2008;3, e2755.
Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, et al. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol. 2010;28:1115–21.
Acknowledgments
D. Bebbere is the recipient of an RTD contract at the University of Sassari, Italy, granted by “P.O.R. SARDEGNA F.S.E. 2007–2013 - Obiettivo competitività regionale e occupazione, Asse IV Capitale umano, Linea di Attività l.3.1.”
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Capsule The present mini review provides an overview of recent findings related to the SCMC and discusses its potential physiological role/s with the aim of inspiring new directions for future research.
Rights and permissions
About this article
Cite this article
Bebbere, D., Masala, L., Albertini, D.F. et al. The subcortical maternal complex: multiple functions for one biological structure?. J Assist Reprod Genet 33, 1431–1438 (2016). https://doi.org/10.1007/s10815-016-0788-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-016-0788-z