Abstract
Purpose
Characterization of the human microbiome has become more precise with the application of powerful molecular tools utilizing the unique 16S ribosomal subunit’s hypervariable regions to greatly increase sensitivity. The microbiome of the lower genital tract can prognosticate obstetrical outcome while the upper reproductive tract remains poorly characterized. Here, the endometrial microbiome at the time of single embryo transfer (SET) is characterized by reproductive outcome.
Methods
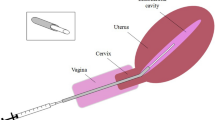
Consecutive patients undergoing euploid, SET was included in the analysis. After embryo transfer, performed as per routine, the most distal 5-mm portion of the transfer catheter was sterilely placed in a DNA free PCR tube. Next-generation sequencing of the bacteria specific 16S ribosome gene was performed, allowing genus and species calls for microorganisms.
Results
Taxonomy assignments were made on 35 samples from 33 patients and 2 Escherichia coli controls. Of the 33 patients, 18 had ongoing pregnancies and 15 did not. There were a total of 278 different genus calls present across patient samples. The microbiome at time of transfer for those patients with ongoing pregnancy vs. those without ongoing pregnancy was characterized by top genera by sum fraction. Lactobacillus was the top species call for both outcomes.
Conclusions
The data presented here show the microbiome at the time of embryo transfer can successfully be characterized without altering standard clinical practice. This novel approach, both in specimen collection and analysis, is the first step toward the goal of determining physiologic from pathophysiologic microbiota. Further studies will help delineate if differences in the microbiome at the time of embryo transfer have a reliable impact on pregnancy outcome.




Similar content being viewed by others
References
Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al. The NIH human microbiome project. Genome Res. 2009;19(12):2317–23.
Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291(5507):1304–51.
Davies J. In a map for human life, count the microbes, too. Science. 2001;291(5512):2316.
Relman DA, Falkow S. The meaning and impact of the human genome sequence for microbiology. Trends Microbiol. 2001;9(5):206–8.
Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345(6270):60–3.
Dymock D, Weightman AJ, Scully C, Wade WG. Molecular analysis of microflora associated with dentoalveolar abscesses. J Clin Microbiol. 1996;34(3):537–42.
Verhelst R, Verstraelen H, Claeys G, Verschraegen G, Delanghe J, Van Simaey L, et al. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis and bacterial vaginosis. BMC Microbiol. 2004;4:16.
Zhou X, Bent SJ, Schneider MG, Davis CC, Islam MR, Forney LJ. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiol Read Engl. 2004;150(Pt 8):2565–73.
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–8.
Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. Microbes on the human vaginal epithelium. Proc Natl Acad Sci U S A. 2005;102(22):7952–7.
Selman H, Mariani M, Barnocchi N, Mencacci A, Bistoni F, Arena S, et al. Examination of bacterial contamination at the time of embryo transfer, and its impact on the IVF/pregnancy outcome. J Assist Reprod Genet. 2007;24(9):395–9.
Hyman RW, Herndon CN, Jiang H, Palm C, Fukushima M, Bernstein D, et al. The dynamics of the vaginal microbiome during infertility therapy with in vitro fertilization-embryo transfer. J Assist Reprod Genet. 2012;29(2):105–15.
Møller BR, Kristiansen FV, Thorsen P, Frost L, Mogensen SC. Sterility of the uterine cavity. Acta Obstet Gynecol Scand. 1995;74(3):216–9.
Mitchell CM, Haick A, Nkwopara E, Garcia R, Rendi M, Agnew K, et al. Colonization of the upper genital tract by vaginal bacterial species in nonpregnant women. Am J Obstet Gynecol. 2015;212(5):611.e1–9.
Swidsinski A, Verstraelen H, Loening-Baucke V, Swidsinski S, Mendling W, Halwani Z. Presence of a polymicrobial endometrial biofilm in patients with bacterial vaginosis. PLoS One. 2013;8(1), e53997.
Treff NR, Ferry KM, Zhao T, Su J, Forman EJ, Scott RT. Cleavage stage embryo biopsy significantly impairs embryonic reproductive potential while blastocyst biopsy does not: a novel paired analysis of cotransferred biopsied and non-biopsied sibling embryos. Fertil Steril. 2011;96:S2.
Treff N, Tao X, Su J, Northrop LE, Kamani M, Bergh P, et al. SNP microarray based concurrent screening of 24 chromosome aneuploidy, unbalanced translocations, and single gene disorders in human embryos: first application of comprehensive triple factor PGD. Biol Reprod. 2009;81:188.
Treff NR, Tao X, Lonczak A, Su J, Taylor D, Scott R. Four hour 24 chromosome aneuploidy screening using high throughput PCR SNP allele ratio analyses. Fertil Steril. 2009;92:S49–50.
Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–7.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinforma Oxf Engl. 2010;26(19):2460–1.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–72.
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–8.
R Development Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing; 2009.
Holmes B, Owen RJ, Steigerwalt AG, Brenner DJ. Flavobacterium gleum, a new species found in human clinical specimens. Int J Syst Evol Microbiol. 1984;34(1):21–5.
Robertson SA, Chin PY, Glynn DJ, Thompson JG. Peri-conceptual cytokines—setting the trajectory for embryo implantation, pregnancy and beyond. Am J Reprod Immunol. 2011;66:2–10.
Soboll G, Shen L, Wira CR. Expression of Toll-like receptors (TLR) and responsiveness to TLR agonists by polarized mouse uterine epithelial cells in culture. Biol Reprod. 2006;75(1):131–9.
Herbst-Kralovetz MM, Quayle AJ, Ficarra M, Greene S, Rose WA, Chesson R, et al. Original article: quantification and comparison of Toll-like receptor expression and responsiveness in primary and immortalized human female lower genital tract epithelia. Am J Reprod Immunol. 2008;59(3):212–24.
Schaefer TM, Fahey JV, Wright JA, Wira CR. Innate immunity in the human female reproductive tract: antiviral response of uterine epithelial cells to the TLR3 agonist poly(I:C). J Immunol Baltim Md 1950. 2005;174(2):992–1002.
O’Neill LAJ. How Toll-like receptors signal: what we know and what we don’t know. Curr Opin Immunol. 2006;18(1):3–9.
Author information
Authors and Affiliations
Corresponding author
Additional information
Capsule The microbiome at the time of embryo transfer can successfully be characterized without altering standard clinical practice utilizing next-generation sequencing of the 16S ribosomal subunit.
Rights and permissions
About this article
Cite this article
Franasiak, J.M., Werner, M.D., Juneau, C.R. et al. Endometrial microbiome at the time of embryo transfer: next-generation sequencing of the 16S ribosomal subunit. J Assist Reprod Genet 33, 129–136 (2016). https://doi.org/10.1007/s10815-015-0614-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-015-0614-z