Abstract
Purpose
Histone H3 lysine 9 (H3K9) methylation plays an important role in the regulation of preimplantation embryo development. G9a has been reported to be a major H3K9mono (m1)/dimethylation(m2) methyltransferase and to contain nuclear localization signals. This study was performed to investigate the correlation between H3K9 methylation level and G9a localization when the nuclear membrane undergoes periodic reconstruction in the cell cycle during preimplantation embryo development.
Methods
The fluorescence intensity was examined via immunofluorescence. The mRNA expression of G9awas determined using real-time reverse transcriptase (RT)-PCR. Eight-cell embryos were cultured in KSOM supplemented with nocodazole (0.5 μM) for 12 h.
Results
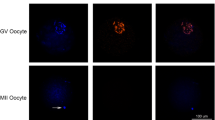
In this study, it was observed that the fluorescence intensity of H3K9m2 and G9a began to increase significantly from the 4-cell stage and reached the peak at the morula stage (p < 0.001), but the fluorescence intensity declined to 4-cell-stage levels when it reached the blastula stage. We observed a similar pattern when we examined G9a mRNA expression. Once the nuclear membrane disintegrated, G9a and H3K9m1 were not detectable by immunofluorescence; when it was reconstructed, G9a and H3K9m1 had relocated to the cell nucleus. However, no significant change was observed in the H3K9m2 localization or in the G9a mRNA level (p > 0.05) during the whole process. JHDM2A was consistently localized in the cytoplasm irrespective of the presence or absence of a nuclear membrane.
Conclusion
These results indicate dynamic changes in the expression level of H3K9m2 and G9a as preimplantation embryogenesis progresses. G9a co-localized with H3K9 m1 in a nuclear membrane-dependent manner during mouse preimplantation embryo development.





Similar content being viewed by others
References
Strahl B, Allis C. The language of covalent histone modifications. Nature. 2000;403:41–5.
Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genes Dev. 2005;15:163–76.
Loh YH, Zhang WW, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–57.
Cabot R. Chromatin remodeling and embryo development. Biol Reprod. 2010;83:98.
Pickard B, Dean W, Engemann S, Bergmann K, Fuermann M, Jung M, Reis A, Allen N, Piotrowska K, Modliński JA, Korwin-Kossakowski M, Karasiewicz J. Effects of preactivation of ooplasts or synchronization of blastomere nuclei in G1 on preimplantation development of rabbit serial nuclear transfer embryos. Biol Reprod. 2000;63:677–82.
Wu SC, Zhang Y. Active DNA demethylation: many roads lead to rome. Nat Rev Mol Cell Biol. 2010;9:607–20.
Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–17.
Estève PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Gene Dev. 2006;20:3089–103.
Myant K, Termanis A, Sundaram AY, Boe T, Li C, Merusi C, Burrage J, de Las Heras JI, Stancheva I. LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res. 2011;21:83–94.
Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, Rossi F, Lorincz MC. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. PNAS. 2011;108:5718–23.
Collins RE, Tachibana M, Tamaru H, Smith KM, Jia D, Zhang X, Selker EU, Shinkai Y, Cheng X. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J Biol Chem. 2005;280:5563–70.
Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91.
Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89.
Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM. G9a selectively represses a class of late-replicating genes at the nuclear periphery. PNAS. 2009;106:19363–8.
Estève PO, Patnaik D, Chin HG, Benner J, Teitell MA, Pradhan S. Functional analysis of the N-and C-terminus of mammalian G9a histone H3 methyltransferase. Nucleic Acids Res. 2005;33:3211–23.
Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–95.
Doherty AS, Mann MRW, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62:1526–35.
Otaegui PJ, O’Neill GT, Campbell KH, Wilmut I. Transfer of nuclei from 8-cell stage mouse embryos following use of nocodazole to control the cell cycle. Mol Reprod Dev. 1994;39:147–52.
Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterize the first cell cycle in mouse embryos. Dev Biol. 2005;280:225–36.
Samake S, Smith LC. Effects of cell-cycle-arrest agents on cleavage and development of mouse embryos. J Exp Zool. 1996;274:111–20.
Matsui Y, Nakayama Y, Okamoto M, Fukumoto Y, Yamaguchi N. Enrichment of cell populations in metaphase, anaphase, and telophase by synchronization using nocodazole and blebbistatin: a novel method suitable for examining dynamic changes in proteins during mitotic progression. Eur J Cell Biol. 2012;91:413–9.
Sun QY, Wu GM, Lai L, Park KW, Cabot R, Cheong HT, Day BN, Prather RS, Schatten H. Translocation of active mitochondria during pig oocyte maturation, fertilization and early embryo development in vitro. Reproduction. 2001;122:155–63.
Korfiatis N, Trounson A, Lacham-Kaplan O. Cell synchronization for the purposes of nuclear transfer in the bovine. Cloning Stem Cells. 2001;3:125–38.
Zhang LS, Jiang MX, Lei ZL, Li RC, Sang D, Sun QY, Chen DY. Development of goat embryos reconstituted with somatic cells: the effect of cell-cycle coordination between transferred nucleus and recipient oocytes. J Reprod Dev. 2004;50:661–6.
Danilchik MV, Bedrick SD, Brown EE, Ray K. Furrow microtubules and localized exocytosis in cleaving Xenopuslaevis embryos. J Cell Sci. 2003;116:273–83.
Hoebeke JC, Van Nijen G, De Brabander M. Interaction of nocodazole(R17934), a new anti-tumoral drug, with rat brain tubulin. Biochem Biophys Res Commun. 1976;69:319–24.
Maro B, Bornens M. The centriole-nucleus association: effects of cytochalasin B and nocodazole. Biol Cells. 1980;39:287–90.
Johnson MH, Pickering SJ, Dhiman A, Radcliffe GS, Maro B. Cytocortical organization during natural and prolonged mitosis of mouse 8-cell blastomeres. Development. 1988;102:143–55.
Sun F, Betzendahl I, Pacchierotti F, Ranaldi R, Smitz J, Cortvrindt R, Eichenlaub-Ritter U. Aneuploidy in mouse metaphase II oocytes exposed in vivo and in vitro in preantral follicle culture to nocodazole. Mutagenesis. 2005;20:65–75.
Yu Y, Xia P, Li S, Yan Y, Tan J. Determination and synchronisation of G1-phase of the cell cycle in 2-and 4-cell mouse embryos. Zygote. 2002;10:245–51.
Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–96.
Zeng F, Schultz RM. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol. 2005;283:40–57.
Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14:47–58.
Kato Y, Ushijima Y, Yamaguchi M. Identification of nuclear localization signals of Drosophila G9a histone H3 methyltransferase. Cell Struct Funct. 2011;36:121–9.
Weiss SL, Lee EA, Diamond J. Evolutionary matches of enzyme and transporter capacities to dietary substrate loads in the intestinal brush border. PNAS. 1998;95:2117–21.
Diamond JM. In: Noble D, Boyd CAR, editors. Logic of life: the challenge of integrative physiology. Oxford: Oxford Univ. Press; 1993. p. 89–111.
Diamond J, Hammond K. The matches, achieved by natural selection, between biological capacities and their natural loads. Experientia. 1992;48:551–7.
Tachibana M, Nozaki M, Takeda N, Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 2007;26:3346–59.
Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–6.
Acknowledgments
This work is supported by the National Key Basic Research (973) program of China (Grant No. 2007CB948101) and Scientific and Technological Developing Scheme of Shaanxi Province (Grant No. 2012K17-02-03). We thank Dr. Lei Pan from Institute of Genetics and Developmental Biology, Chinese Academy of Sciences for skillful technical assistance. We also appreciate the valuable comments from other members of our laboratory.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Capsule
G9a specifically regulates histone H3 lysine 9 monomethylation in a nuclear membrane-dependent manner during mouse preimplantation embryo development.
Bo Li and Na Tang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Li, B., Tang, N., Chen, S. et al. G9a co-localized with histone H3 lysine 9 monomethylation but not dimethylation in a nuclear membrane-dependent manner during mouse preimplantation embryo development. J Assist Reprod Genet 30, 441–448 (2013). https://doi.org/10.1007/s10815-012-9911-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-012-9911-y