Abstract
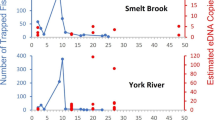
Environmental DNA (eDNA) studies show great promise for non-invasive surveys of aquatic organisms, but should account for imperfect detection and the influences of biotic and abiotic conditions on detection. We evaluated an eDNA protocol for Roanoke logperch (RLP) Percina rex, an endangered fish of the eastern United States occupying habitats ranging from cold, clear creeks to warm, turbid rivers. We assayed water samples from streams presumed likely to be occupied or unoccupied by RLP based on previous fish surveys. Data were analyzed using multi-scale occupancy models that estimated occurrence and detection probability at the scales of sites, replicate water filters, and replicate PCR reactions, and environmental influences on these probabilities. We detected RLP eDNA at 11 of 12 sites in occupied streams and no sites in presumed-unoccupied streams. In best-supported models, detection was positively related to an index of fish density, whereas other environmental factors had no consistent effects. This approach had a higher detection rate and lower sensitivity to sampling conditions than traditional techniques like snorkeling and electrofishing, suggesting it could provide a powerful tool for assessing the distribution of this and other rare fishes that occur across a wide range of fluvial habitats.


Similar content being viewed by others
References
Amberg, J. J., S. G. McCalla, E. Monroe, R. Lance, K. Baerwaldt & M. P. Gaikowsk, 2015. Improving efficiency and reliability of environmental DNA analysis for silver carp. Journal of Great Lakes Research 41: 367–373.
Baldigo, B. P., L. A. Sporn, S. D. George & J. A. Ball, 2017. Efficacy of environmental DNA to detect and quantify brook trout populations in headwater streams of the Adirondack Mountains, New York. Transactions of the American Fisheries Society 146: 99–111.
Barnes, M. A., C. R. Turner, C. L. Jerde, M. A. Renshaw, W. L. Chadderton & D. M. Lodge, 2014. Environmental conditions influence eDNA persistence in aquatic systems. Environmental Science and Technology 48: 1819–1827.
Boothroyd, M., N. E. Mandrak, M. Fox & C. C. Wilson, 2016. Environmental DNA (eDNA) detection and habitat occupancy of threatened spotted gar (Lepisosteus oculatus). Aquatic Conservation 26: 1107–1119.
Buxton, A. S., J. J. Groombridge & R. A. Griffiths, 2018. Seasonal variation in environmental DNA detection in sediment and water samples. PLoS ONE 13: e0191737.
Deiner, K. & F. Altermatt, 2014. Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE 9(2): 88786.
Dejean, T., A. Valentini, A. Duparc, S. Pellier-Cuit, F. Pompanon, P. Taberlet & C. Miaud, 2011. Persistence of environmental DNA in freshwater ecosystems. PLoS ONE 6: e23398.
Doi, H., R. Inui, Y. Akamatsu, K. Kanno, H. Yamanaka, T. Takahara & T. Minamoto, 2017. Environmental DNA analysis for estimating the abundance and biomass of stream fish. Freshwater Biology 62: 30–39.
Ellison, S. R., C. A. English, M. J. Burns & J. T. Keer, 2006. Routes to improving the reliability of low level DNA analysis using real-time PCR. BMC Biotechnology 6: 33–43.
Furlan, E. M., D. Gleeson, C. M. Hardy & R. P. Duncan, 2016. A framework for estimating the sensitivity of eDNA surveys. Molecular Ecology Resources 16: 641–654.
Goldberg, C. S., C. R. Turner, K. Deiner, K. E. Klymus, P. F. Thomsen, M. A. Murphy, S. F. Spear, A. McKee, S. J. Oyler-McCance, R. S. Cornman, M. B. Laramie, A. R. Mahon, R. F. Lance, D. S. Pilliod, K. M. Strickler, L. P. Waits, A. K. Fremier, T. Takahara, J. E. Herder & P. Taberlet, 2016. Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods in Ecology and Evolution 7: 1299–1307.
Hines, J. E., 2006. PRESENCE: Software to estimate patch occupancy and related parameters. United States Geological Survey, Patuxent Wildlife Research Center. [available on internet at http://www.mbr-pwrc.usgs.gov/software/presence.html.
Hunter, M. E., S. J. Oyler-McCance, R. M. Dorazio, J. A. Fike, B. J. Smith, C. T. Hunter & K. M. Hart, 2015. Environmental DNA (eDNA) sampling improves occurrence and detection estimates of invasive burmese pythons. PLoS ONE 10: e0121655.
Jane, S. F., T. M. Wilcox, K. S. McKelvey, M. K. Young, M. K. Schwartz, W. H. Lowe, B. H. Letcher & A. R. Whiteley, 2015. Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Molecular Ecology Resources 15: 216–227.
Janosik, A. M. & C. E. Johnston, 2015. Environmental DNA as an effective tool for detection of imperiled fishes. Environmental Biology of Fishes 98: 1889–1893.
Jerde, C. L., A. R. Mahon, W. L. Chadderton & D. M. Lodge, 2011. “Sight-unseen” detection of rare species using environmental DNA. Conservation Letters 4: 150–157.
Jerde, C. L., B. P. Olds, A. J. Shogren, E. A. Andruszkiewicz, A. R. Mahon, D. Bolster & J. L. Tank, 2016. Influence of stream bottom substrate on retention and transport of vertebrate environmental DNA. Environmental Science and Technology 50(16): 8770–8779.
Laramie, M. B., D. S. Pilliod & C. S. Goldberg, 2015. Characterizing the distribution of an endangered salmonid using environmental DNA analysis. Biological Conservation 183: 29–37.
MacKenzie, D. I., J. D. Nichols, J. A. Royle, K. H. Pollock, L. L. Bailey & J. E. Hines, 2006. Occupancy estimation and modeling: Inferring patterns and dynamics of species occurrence. Academic Press, Burlington.
Maruyama, A., K. Nakamura, H. Yamanaka, M. Kondoh & T. Minamoto, 2014. The release rate of environmental DNA from juvenile and adult fish. PLoS ONE 10: e0118727.
Matthews, W. J., 1998. Patterns in freshwater fish ecology. Chapman and Hall, New York.
McKee, A. M., S. F. Spear & T. W. Pierson, 2015. Special Issue Article: Environmental DNA: The effect of dilution and the use of a post-extraction nucleic acid purification column on the accuracy, precision, and inhibition of environmental DNA samples. Biological Conservation 183: 70–76.
Moyer, G. R., E. Díaz-Ferguson, J. E. Hill & C. Shea, 2014. Assessing Environmental DNA Detection in Controlled Lentic Systems. PLoS ONE 9: e103767.
Nevers, M. B., M. N. Byappanahalli, C. C. Morris, D. Shively, K. Przybyla-Kelly, A. M. Spoljaric & E. F. Roseman, 2018. Environmental DNA (eDNA): A tool for quantifying the abundant but elusive round goby (Neogobius melanostomus). PLoS ONE 9: e0191720.
Nichols, J. D., L. L. Baile, N. W. Talancy, E. H. Campbell Grant, A. T. Gilbert, E. M. Annand, T. P. Husband & J. E. Hines, 2008. Multi-scale occupancy estimation and modelling using multiple detection methods. Journal of Applied Ecology 45: 1321–1329.
Pavlacky, D. C., J. A. Blakesley, G. C. White, D. J. Hanni & P. M. Lukacs, 2012. Hierarchical multi-scale occupancy estimation for monitoring wildlife populations. The Journal of Wildlife Management 76: 154–162.
Peterson, J. T. & C. P. Paukert, 2009. Converting nonstandard fish sampling data to standardized data. In Bonar, S. A., W. A. Hubert & D. W. Willis (eds), Standard Methods for Sampling North American Freshwater Fishes. American Fisheries Society, Bethesda: 195–215.
Pfleger, M. O., S. J. Rider, C. E. Johnston & A. M. Janosik, 2016. Saving the doomed: Using eDNA to aid in detection of rare sturgeon for conservation (Acipenseridae). Global Ecology and Conservation 8: 99–107.
Pilliod, D. S., C. S. Goldberg, R. S. Arkle & L. P. Waits, 2013a. Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Canadian Journal of Fisheries and Aquatic Sciences 70: 1123–1130.
Pilliod, D. S., C. S. Goldberg, R. S. Arkle & L. P. Waits, 2013b. Factors influencing detection of eDNA from a stream-dwelling amphibian. Molecular Ecology Resources 14: 109–116.
Pont, D., M. Rocle, A. Valentini, R. Civade, P. Jean, A. Maire, N. Roset, M. Schabuss, H. Zornig & T. Dejean, 2018. Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Scientific Reports 8: 10361.
Rabeni, C. F., J. Lyons, N. Mercado-Silva & J. T. Peterson, 2009. Warmwater fish in wadeable streams. In Bonar, S. A., W. A. Hubert & D. W. Willis (eds), Standard methods for sampling North American freshwater fishes. American Fisheries Society, Bethesda: 43–58.
Renshaw, M. A., B. P. Olds, C. L. Jerde, M. M. McVeigh & D. M. Lodge, 2015. The room temperature preservation of filtered environmental DNA samples and assimilation into a phenol–chloroform–isoamyl alcohol DNA extraction. Molecular Ecology Resources 15: 168–176.
Roberts, J. H., 2012. Assessment of the distribution and abundance of Roanoke logperch (Percina rex) in the Dan River basin of Virginia. Final Report to Virginia Department of Game and Inland Fisheries, Richmond.
Roberts, J. H., P. L. Angermeier & E. M. Hallerman, 2013. Distance, dams, and drift: what structures populations of an endangered, benthic stream fish? Freshwater Biology 58: 2050–2064.
Robson, H. L. A., T. H. Noble, R. J. Saunders, S. K. A. Robson, D. W. Burrows & D. R. Jerry, 2016. Fine-tuning for the tropics: Application of eDNA technology for invasive fish detection in tropical freshwater ecosystems. Molecular Ecology Resources 16(4): 922–932.
Rosenberger, A. E., 2007. An update to the Roanoke logperch recovery plan. Final Report to the U.S. Fish and Wildlife Service, Gloucester, Virginia.
Schmelzle, M. C. & A. P. Kinziger, 2016. Using occupancy modelling to compare environmental DNA to traditional field methods for regional-scale monitoring of an endangered aquatic species. Molecular Ecology Resources 16: 895–908.
Schmidt, B. R., M. Kéry, S. Ursenbacher, O. J. Hyman, J. P. Collins & N. Yoccoz, 2013. Site occupancy models in the analysis of environmental DNA presence/absence surveys: a case study of an emerging amphibian pathogen. Methods in Ecology & Evolution 4: 646–653.
Schrader, C., A. Schielke, L. Ellerbroek & R. Johne, 2012. PCR inhibitors—occurrence, properties and removal. Journal of Applied Microbiology 113: 1014–1026.
Schultz, M. T. & R. F. Lance, 2015. Modeling the sensitivity of field surveys for detection of environmental DNA (eDNA). PLoS ONE 10(10): e0141503.
Shogren, A. J., J. L. Tank, E. Andruszkiewicz, B. Olds, A. R. Mahon, C. L. Jerde & D. Bolster, 2017. Controls on eDNA movement in streams: transport, retention, and resuspension. Scientific Reports 7: 5065.
Stoeckle, B. C., S. Beggel, A. F. Cerwenka, E. Motivans, R. Kuehn, J. Geist & H. Doi, 2017. A systematic approach to evaluate the influence of environmental conditions on eDNA detection success in aquatic ecosystems. PLOS ONE 12(12): e0189119.
Strickler, K. M., A. K. Fremier & C. S. Goldberg, 2015. Special Issue Article: Environmental DNA: Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biological Conservation 183: 85–92.
Takahara, T., T. Minamoto, H. Yamanaka, H. Doi & Z. Kawabata, 2012. Estimation of fish biomass using environmental DNA. PLoS ONE 7: e35868.
Takahara, T., T. Minamoto & H. Doi, 2013. Using environmental DNA to estimate the distribution of an invasive fish species in ponds. PLoS ONE 8: e56584.
Tamura, K., G. Stecher, D. Peterson, A. Filipski & S. Kumar, 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729.
Thomsen, P. F. & E. Willerslev, 2015. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biological Conservation 183: 4–18.
Thomsen, P. F., J. O. S. Kielgast, L. L. Iversen, C. Wiuf, M. Rasmussen, M. T. P. Gilbert, L. Orlando & E. Willerslev, 2012. Monitoring endangered freshwater biodiversity using environmental DNA. Molecular Ecology 21: 2565–2573.
Tillotson, M. D., R. P. Kelly, J. J. Duda, M. Hoy, J. Kralj & T. P. Quinn, 2018. Concentrations of environmental DNA (eDNA) reflect spawning salmon abundance at fine spatial and temporal scales. Biological Conservation 220: 1–11.
Turner, C. R., D. J. Miller, K. J. Coyne & J. Corush, 2014a. Improved methods for capture, extraction, and quantitative assay of environmental DNA from Asian bigheaded carp (Hypophthalmichthys spp.). PloS ONE 9: e114329.
Turner, C. R., K. L. Uy & R. C. Everhart, 2015. Fish environmental DNA is more concentrated in aquatic sediments than surface water. Biological Conservation 183: 93–102.
Turner, C. R., M. A. Barnes, C. C. Y. Xu, S. E. Jones, C. L. Jerde & D. M. Lodge, 2014b. Particle size distribution and optimal capture of aqueous macrobial eDNA. Methods in Ecology and Evolution 5: 676–684.
Wang, Y., 2004. A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Research 32(3): 1197–1207.
Wilcox, T. M., K. S. McKelvey, M. K. Young, S. F. Jane, W. H. Lowe, A. R. Whiteley & M. K. Schwartz, 2013. Robust detection of rare species using environmental DNA: The importance of primer specificity. PLoS ONE 8: e59520.
Wilcox, T. M., K. S. McKelvey, M. K. Young, W. H. Lowe, M. K. Schwartz, 2015. Environmental DNA particle size distribution from Brook Trout (Salvelinus fontinalis). Conservation Genetics Resources 7(3): 639–641.
Wilcox, T. M., K. S. McKelvey, M. K. Young, A. J. Sepulveda, B. B. Shepard, S. F. Jane, A. R. Whiteley, W. H. Lowe & M. K. Schwartz, 2016. Understanding environmental DNA detection probabilities: A case study using a stream-dwelling char Salvelinus fontinalis. Ecological Applications 194: 209–216.
Williams, K. E., K. P. Huyvaert & A. J. Piaggio, 2017. Clearing muddied waters: Capture of environmental DNA from turbid waters. PLoS ONE 12: e0179282.
Willoughby, J. R., B. K. Wijayawardena, M. Sundaram, R. K. Swihart & J. A. DeWoody, 2016. The importance of including imperfect detection models in eDNA experimental design. Molecular Ecology Resources 16: 837–844.
Ye, J., G. Coulouris, I. Zaretskaya, I. Cutcutache, S. Rozen & T. L. Madden, 2012. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 1–11.
Acknowledgements
This project was funded by the Virginia Department of Game and Inland Fisheries (Fund #2014-14502). We thank M. Pinder for his assistance in bringing it to fruition. M. Moore, G. Moyer, T. Darden, M. Walker, and C. Cutler kindly helped us troubleshoot technical issues. J. Eschenroeder and B. Blood assisted with the creation of figures. This study was funded by the Virginia Department of Game and Inland Fisheries.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Eric Larson
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Strickland, G.J., Roberts, J.H. Utility of eDNA and occupancy models for monitoring an endangered fish across diverse riverine habitats. Hydrobiologia 826, 129–144 (2019). https://doi.org/10.1007/s10750-018-3723-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-018-3723-8