Abstract
Hybridization between introduced and indigenous species can lead to loss of unique genetic resources and precipitate extinction. In Tanzania, the Nile tilapia (Oreochromis niloticus) and blue-spotted tilapia (Oreochromis leucostictus) have been widely introduced to non-native habitats for aquaculture and development of capture fisheries. Here, we aimed to quantify interspecific hybridization between these introduced species and the indigenous species Oreochromis esculentus, Oreochromis jipe and Oreochromis korogwe. In the Pangani basin, several hybrids were observed (O. niloticus × O. jipe, O. leucostictus × O. jipe, O. niloticus × O. korogwe), although hybrids were relatively uncommon within samples relative to purebreds. Hybrids between the native O. jipe × O. korogwe were also observed. In the Lake Victoria basin, no evidence of hybrids was found. Analysis of body shape using geometric morphometrics suggested that although purebreds could be discriminated from one another, hybrids could not be readily identified on body and head shape alone. These results provide the first evidence of hybridization between the introduced species and the Critically Endangered O. jipe in Tanzania. Given uncertainty regarding benefits of introduced species over large-bodied indigenous species in aquaculture and capture fisheries, we suggest that future introductions of hybridization-prone species should be carefully evaluated.
Similar content being viewed by others
Introduction
Introduced species are recognized as a major driver of biodiversity loss in aquatic environments (Clavero & García-Berthou, 2005), and many aquatic introduced species have been associated with substantial economic and ecological impacts (Lowe et al., 2000; Pimentel et al., 2005; Lowell et al., 2006). The spread of these species is predicted to continue as natural biogeographic barriers are overcome, either accidentally through unintended transport, or through deliberate introductions (Levine & D’Antonio, 2003; Taylor & Irwin 2004; Hulme et al., 2008; Westphal et al., 2008). Globally, there are few regions that have not been invaded by at least one introduced aquatic species (Molner et al., 2008; Leprieur et al., 2008), with global hotspots being highly correlated with human activity and trade (Drake & Lodge, 2003; Perrings et al., 2005; Meyerson & Mooney, 2007). Freshwater environments are considered especially vulnerable to invasion (Sala et al., 2000; Cox & Lima, 2006). A key concern is the inability to reverse invasions and the subsequent impacts, and only a few species have ever been successfully eradicated from an aquatic environment after establishment (Hill & Cichra, 2005; Williams & Grosholz, 2008; Leprieur et al., 2009; Hill & Sowards, 2015).
The range of ecological impacts associated with introduced species is extensive, including predation, competition and habitat alteration (Canonico et al., 2005). There is also growing concern surrounding the threat of the loss of indigenous unique genetic diversity through hybridization between native and introduced species, potentially even to the extent of species extinction (Levin et al., 1996; Rhymer & Simberloff, 1996). Hybridization is of particular concern in cases where one of the species are considered to be a threatened species. For example, in North America, vulnerable endemic Pecos pupfish Cyprinodon pecosensis (Echelle & Echelle 1978) have hybridized with the invasive sheepshead minnow Cyprinodon variegatus (Lacepède 1803) with potentially no pure populations remaining due to the apparent vigour of hybrid individuals (Rosenfield et al., 2004).
Hybridization has been invoked as a potential driver of biodiversity loss in the tilapiine cichlid fish of the genus Oreochromis. Several species have been introduced to non-native habitats in Africa with a view to developing and improving capture fisheries and aquaculture. Among the most widely distributed species is the Nile tilapia (Oreochromis niloticus L. 1758), native to the Nile basin and West Africa (Trewavas, 1983), but introduced into at least 15 African countries outside its native range (FAO, 2012). Oreochromis species are known for interspecific hybridization (Scribner et al., 2001), and hybrids are commonplace in aquaculture where they are selected for desirable characteristics such as salinity tolerance (Agresti et al., 2000; Kamal & Mair, 2005) and pigmentation (McAndrew et al., 1988; Romana-Eugia et al., 2004). In the natural environment, introduced O. niloticus has been documented as hybridizing with several species including Oreochromis mossambicus (Peters 1852) in South Africa (D’Amato et al., 2007), Oreochromis aureus (Steindachner 1864) in Egypt and West Africa (Rognon & Guymard, 2003; Bakhoum et al., 2009), Oreochromis andersonii (Castelnau 1861) and Oreochromis macrochir (Boulenger 1912) in Zambia (Deines et al., 2014), and Oreochromis esculentus (Graham 1928) and Oreochromis leucostictus (Trewavas 1933) in Kenya (Nyingi & Agnèse 2007; Angienda et al., 2011; Ndiwa et al., 2014). However, the underlying factors mediating levels of hybridization between Oreochromis are poorly understood.
In southern and eastern Africa, O. niloticus is now present in multiple major drainage systems where it was historically absent, including the Pangani, Rufiji, Ruvuma, Limpopo, Zambezi and Lake Victoria basins (Genner et al., 2013; Zengeya et al., 2013; Deines et al., 2014; Shechonge et al. unpublished data). This spread of O. niloticus has been accompanied by blue-spotted tilapia (O. leucostictus), native to Lakes Edward, George and Albert in the Nile system (Trewavas 1983; Shechonge et al. unpublished data). The consequences of introducing these species for native fauna in their new range are largely unknown (Deines et al., 2016). Concerns exist for both the conservation of native endangered populations (Gregg et al., 1998; Moralee et al., 2000; Nyingi & Agnèse, 2007; Angienda et al., 2011) as well as the preservation of wild genetic resources for aquaculture purposes (Lind et al., 2012).
In this study, we focus on the extent of hybridization between native and introduced Oreochromis in the Lake Victoria and Pangani basins of northern Tanzania. Both basins are characterized by endemic species that are threatened by negative interactions, including hybridization with introduced O. niloticus and O. leucostictus. Specifically, in the Lake Victoria basin, the Critically Endangered endemic O. esculentus persists only in satellite water bodies of Lake Victoria, although it has been translocated to multiple locations in surrounding countries (Trewavas, 1983; Shechonge et al. unpublished data). In the Pangani system, the Critically Endangered endemic Oreochromis jipe (Lowe 1955) is distributed from Lake Jipe to the Pangani Falls dam where it co-occurs with the lowland native species Oreochromis korogwe (Lowe 1955) (Trewavas, 1983; Shechonge et al. unpublished data). Given an apparent threat to Critically Endangered species, here we assess the extent of hybridization with introduced Oreochromis species using genetic (microsatellite) markers. We also asked if genetically identified hybrids could be identified using geometric morphometric data that capture variation in external head and body shape, potentially informing future field survey work on hybrid abundance.
Methods
Sampling
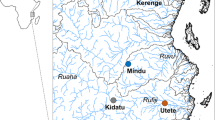
We collected our focal Oreochromis samples from one site in the Lake Victoria catchment and four sites in the Pangani river drainage (Fig. 1; Table 1). Sampling took place in 2015 and 2016 using seine nets, gill nets and purchasing from artisanal fisherman. Samples of additional reference species were obtained from five other sites in Tanzania (Table 1), where the species were found alone, or co-existing with species without any morphological or genetic evidence of hybridization. Fish captured using netting were immediately euthanized on landing by an overdose of clove oil anaesthetic. Specimens were pinned out and imaged in the field prior to preservation in absolute ethanol. Whole fish were placed in 70% ethanol for long-term storage. Genetic samples (fin clips) were taken and preserved in absolute ethanol.
Microsatellite genotyping
DNA was extracted from fin tissues using the Promega Wizard DNA extraction kit. Samples were screened at 17 microsatellite loci (Supporting Information Table 1). PCR was performed in a volume of 10 μl PCR solution consisting of 1 μl DNA (~ 5 ng), 5 μl Mastermix and 4 μl primer mix (10 mM). PCR amplifications were conducted on BioRad MyCycler thermal cycler, with conditions consisting of one denaturation step of 15 min at 95°C, followed by 35 cycles of 30 s at 94°C, 90 s at 57°C and 1 min at 72°C, followed by a final extension step of 30 min at 60°C. PCR products were sized on an ABI 3500 automated sequencer against a LIZ 500 size standard using GeneMapper 4.1 (Applied Biosystems).
Microsatellite data analysis
All individuals were amplified at a minimum of 11 of the 17 microsatellite markers (Supporting Information Tables 2 & 3). Genetic diversity estimates, and tests of deviation from Hardy–Weinberg Equilibrium, were calculated in Arlequin 3.5 (Excoffier & Lischer, 2010). To estimate the genetic composition of individuals within sites, we used a two-step process. First we made an initial assignment of focal and reference individuals to species group using find clusters in the R package adegenet (Jombart & Ahmed, 2011), selecting the maximum number of Principal Components possible, and a K value reflecting the number of species present, based on their phenotypes. We then used this initial assignment to groups as prior (LOCPRIOR) in Structure 2.3.4 (Pritchard et al., 2000), selecting the admixture model, and 10 runs, each with a burn-in of 100,000 steps and 100,000 recorded iterations. Next, Clumpak (Kopelman et al., 2015) was used to summarize the Structure output. Individuals with an assignment probability lower than 0.9 to any one of the focal species were considered of hybrid origin.
Geometric morphometrics
The left-hand side of the specimens was photographed with a scale. Images were loaded into tpsDIG v.2.22 (Rohlf, 2005) and 21 landmarks were digitized (Fig. 2). Landmarks were chosen based on landmarks commonly used in morphometric studies (Genner et al., 2007). Shape was quantified using MorphoJ 1.06 (Klingenberg, 2011). In each analysis, a Procrustes superimposition was applied to landmarked data. To visualize shape differences among purebred individuals at the sites, we used a Canonical Variates Analysis in MorphoJ 1.06. To determine repeatability of landmarking, we landmarked a set of 24 randomly selected individuals a second time. Following Procrustes alignment in MorphoJ 1.06, coordinates were subjected to Procrustes Anova the R package Geomorph (Adams et al., 2017) that revealed 54.85% of the total variance to be among individuals, 0.05% of the total variance between sets (original vs. repeat), and 45.10% representing residual variance.
To determine if hybrid individuals possessed intermediate morphology between parent species, we used a discriminant function analysis on procrustes scores in SPSS v.23.0 (IBM), using stepwise variable removal process that retained only the most informative variables. Genotypically “purebred” individuals were assigned to species groups, while putatively hybrid individuals were left unclassified. Individuals from Lake Kumba and Kerenge were pooled for morphological analyses due to their close geographic proximity and overlapping species composition. Individuals from the other three sites were analysed separately.
Results
Microsatellite results
At each of the five sites, the complete samples showed significantly higher heterozygosity relative to expectations from Hardy–Weinberg equilibrium at most loci (Supporting Information Table 2). Analyses of these data supported the presence of O. niloticus at all five sites (Fig. 3), Lake Malimbe (n = 14), Kerenge (n = 30), Lake Kumba (n = 71), Nyumba-ya-Mungu (n = 14) and Pangani Falls Dam (n = 26). O. leucostictus was resolved as present at two sites, Lake Malimbe (n = 31) and Kerenge (n = 9). O. esculentus was present at two sites, Lake Malimbe (n = 11) and Nyumba-ya-Mungu (n = 3). O. jipe was present at three sites, Lake Kumba (n = 13), Nyumba-ya-Mungu (n = 18) and the Pangani Falls Dam (n = 6). O. korogwe was only found at the Pangani Falls Dam (n = 3). We found no evidence of hybrid individuals within Lake Malimbe or Lake Kumba. Evidence supporting the presence of hybrids was found at three sites (Fig. 3). One hybrid individual was identified at Kerenge sample (O. leucostictus × O. jipe). Two O. niloticus × O. jipe hybrids were in the Nyumba-ya-Mungu sample, and eight hybrid individuals were found in the sample from Pangani Falls (one O. korogwe × O. niloticus, and seven O. korogwe × O. jipe).
Posterior probabilities of assignment by structure to species groups of individuals collected at the five study sites, relative to the putatively purebred reference individuals. Each individual is represented by on vertical bar, with colours representing assignment probabilities to the species group. Reference individuals are underlined in black. Asterisk indicates putatively hybrid individuals
Morphological characterization of purebred and hybrid individuals
Geometric morphometrics demonstrated significant differences in geometric morphometric space among purebred individuals (Table 2). Both CVA and discriminant analysis were consistent with sympatric populations being separable on shape variables (Figs. 4, 5), with evident differences primarily in body depth and eye size among species. In discriminant analyses, individuals identified as genetic hybrids were typically, but not exclusively, within the phenotypic space of parental purebred individuals (Fig. 5).
Canonical variate analysis of geometric morphometric shape variation among individuals identified as purebred. Individuals from Kerenge and Lake Kumba are grouped. Outline diagrams demonstrate the variation along CV axes, with line colours representing the species at each of the extremes of the axis
Discussion
We found no evidence of hybridization between O. esculentus, O. niloticus and O. leucostictus at any sites where pairs of these species co-occurred, including Lake Malimbe in the Lake Victoria catchment. Within the Pangani system, we found evidence for the presence of hybrid individuals of the Critically Endangered O. jipe with introduced species at two locations. At Kerenge one individual O. leucostictus × O. jipe was discovered, while at Nyumba-ya-Mungu two O. niloticus × O. jipe were found. Notably, at all these sites hybrid individuals between introduced and native species were uncommon relative to purebred individuals. The relatively low frequency of hybridization in some of our sites is consistent with evidence from other Oreochromis systems, such as introduced O. leucostictus and native O. niloticus in Kenya (Nyingi & Agnèse, 2007; Ndiwa et al., 2014). These patterns contrast with observations of extensive hybridization between introduced O. niloticus and native O. mossambicus in South Africa (D’Amato et al., 2007), and between introduced O. niloticus and native Oreochromis in Zambia (O. macrochir and O. andersonii; Deines et al., 2014).
The relative rarity of hybrids between introduced and indigenous species can perhaps be explained by strong prezygotic isolating mechanisms. Sexual selection acting on male traits such as breeding colour, courtship displays and spawning “bower” phenotypes have all been suggested to promote reproductive isolation in mouthbrooding cichlids (Seehausen et al., 1997, 2008). Additionally, there is a possibility that populations may mate assortatively due to separate breeding periods or different breeding habitat preference, but at present no information on the habitat choice of these species is available. Postzygotic mechanisms may also have contributed to an apparent absence of hybrid individuals in our samples, if hybrid individuals suffer from low viability or fertility relative to purebred individuals. The extent of viability and fertility among the Oreochromis in our study is uncertain, although we note that hybrids of several Oreochromis species have been generated within aquaculture producing viable and fertile offspring (Bartley et al., 2001). In principle, the extent of inviability will be dependent on the extent of genomic incompatibility linked to the timescale since divergence (Bolnick & Near, 2005; Stelkens et al., 2009). However, the fitness of hybrid fish will also be dependent on the specific phenotypes of hybrids relative to the parental lines within the local selective regime. For example, phenotypically intermediate hybrids of cyprinids (Nilsson et al., 2017) and cichlids (Maan et al., 2017) have been demonstrated to have reduced survival relative to parental forms, while hybrids of two centrarchid species have an inferior feeding performance relative to their parental species (McGee et al., 2015).
The evidence of hybridization between the sympatric native species O. korogwe and O. jipe was notable in the Pangani Falls Dam, constructed in 1994. Surveys have reported these species that are otherwise allopatric in their distributions, with O. korogwe being distributed in low altitude coastal stretches of the Pangani and Zigi rivers, while O. jipe is primarily a higher altitude inland species (Trewavas, 1983; Shechonge et al., unpublished data). Further work is needed to map the distributions of both species within the lower reaches of the Pangani river system. It is possible that there is a natural hybrid zone, but it is possible that hybridization has been promoted by the habitat modification either linked to the dam construction, or the presence of O. niloticus. Dam construction has resulted in hybridization in other freshwater fishes (Hasselman et al., 2014), while introduced species have been suggested to affect the natural reproductive behaviour of native fish species (Doupe et al., 2009), including driving the loss of unique genetic diversity though hybridization (Velema et al., 2012).
Timescale of spread of non-native species
An important factor determining the extent of hybridization and negative ecological effects on native fauna is the timescale of invasion. If hybridization is dependent on the density of the invader, then evidence of hybridization may be absent or rare until the invader becomes established and passes a threshold density, resulting in a lag time between the appearance of the non-native species and production of hybrids (Crooks & Soulé, 1999). Oreochromis niloticus and O. leucostictus are likely to have been first introduced into the Pangani system within the last 40 years. Oreochromis niloticus was notably absent from extensive surveys of Nyumba-ya-Mungu in 1974 (Bailey et al., 1978). At that time, O. esculentus was already established and abundant in the dam and forming an important part of the fishery. Given that O. esculentus had been reared in ponds in the lower Pangani near Korogwe as early as 1950 (Lowe-McConnell, 2006), it is possible that it arrived in the upper Pangani region prior to construction of the Nyumba-ya-Mungu dam in 1967–1969, and expanded in population size due to favourable conditions. Introductions of O. leucostictus and O. niloticus into Lake Victoria took place in 1953 (Pringle, 2005; Lowe-McConnell, 2006), but neither had been recorded in satellite Lake Malimbe as recently January 2003 (Katunzi & Kishe, 2004). Given the relatively recent timeline of the arrival of introduced species, future monitoring of genetic structure in these habitats may provide evidence of how shifts in density and time affect the frequency of hybridization.
Morphological evaluation of hybrids
In natural systems, it can be possible to identify hybrid individuals on the basis of morphological characters. For example, F1 hybrids between the European cyprinids roach Rutilus rutilus (L. 1758) and bream Abramis brama (L. 1758) in Ireland can be identified using geometric morphometric analysis of body shape (Hayden et al., 2010), while hybrids of Atlantic salmon Salmo salar (L. 1758) and brown trout Salmo trutta (L. 1758) also typically exhibit intermediate morphology when measured using geometric morphometric approaches (Solem et al., 2014). We found that it was possible to separate purebred individuals of different species in sympatry using shape information, but hybrids overlapped in morphospace with parental individuals. Thus, we suggest that conclusive assignment of some Oreochromis hybrids may not be possible from geometric morphometric data of the gross body shape alone, although perhaps the method may be useful when used in combination with other phenotypic traits such melanin patterning.
Biodiversity and fisheries implications
Hybridization can result in biodiversity loss through genetic swamping, where hybridization leads to the loss of unique genetic diversity, or demographic swamping, where the numeric increases in hybrids result in negative demographic consequences for the parental species (Todesco et al., 2016). The absence or low frequency of hybrids between introduced and indigenous species suggests neither of these scenarios are likely under current environmental regimes at our study sites. However, hybridization at low frequencies can lead to the introduction of novel alleles that either reduce fitness of native species (Muhlfield et al., 2009), or promote traits such as fast maturation and small body sizes that could compromise fish production. By contrast, hybridization can promote the sharing of beneficial alleles, reducing vulnerability to inbreeding and disease and facilitating increased niche width by both native and introduced populations (Hall, 2016; Pfennig et al., 2016). In principle, this can lead to net benefits to capture fisheries through improving survivorship. In Oreochromis, it is unclear if the presence of hybridization between native and non-native species has affected production in either capture fisheries or aquaculture, but these questions could be investigated through further work on the genomic composition of these Oreochromis communities, together with common-garden experiments investigating traits related to fish production.
Concluding remarks
Fisheries in East Africa are essential for local livelihoods and food security (Muir et al., 2005; Heck et al., 2007; Musaka & Musonda, 2013), and the introduced species O. niloticus and O. leucostictus are now important components of demersal fisheries in northern Tanzania (e.g. Kolding et al., 2014). However, any benefits from further spread of introduced species in East Africa must be weighed against potential risks to biodiversity, existing stocks, and future potential fisheries yields. We propose that fisheries managers adopt the precautionary principle, that suggests future aquaculture and capture fisheries development should be based primarily on indigenous large-bodied species, unless there is compelling evidence that the economic and societal benefits will outweigh risks to biodiversity and existing artisanal fisheries. Further information on the likelihood of hybridization among species under different environmental conditions would help to guide policy and fisheries development in the region.
References
Adams, D. C., M. L. Collyer, A. Kaliontzopoulou, & E. Sherratt. 2017. Geomorph: Software for geometric morphometric analyses. R package version 3.0.5. https://cran.r-project.org/package=geomorph.
Agresti, J. J., S. Seki, A. Cnaani, S. Poompuang, E. M. Hallerman, N. Umiel, G. Hulata, G. A. E. Gall & B. May, 2000. Breeding new strains of tilapia: development of an artificial center of origin and linkage map based on AFLP and microsatellite loci. Aquaculture 185: 43–56.
Angienda, P. O., H. J. Lee, K. R. Elmer, R. Abila, E. N. Waindi & A. Meyer, 2011. Genetic structure and gene flow in an endangered native tilapia fish (Oreochromis esculentus) compared to invasive Nile tilapia (Oreochromis niloticus) in Yala Swamp, East Africa. Conservation Genetics 12: 243–255.
Bailey, R. G., S. Churchfield, T. Petr & R. Pimm, 1978. The ecology of the fishes in Nyumba ya Mungu reservoir, Tanzania. Biological Journal of the Linnean Society 10: 109–137.
Bakhoum, S. A., M. A. Sayed-Ahmed & E. A. Ragheb, 2009. Genetic evidence for natural hybridization between Nile tilapia (Oreochromis niloticus: Linnaeus 1757) and Blue tilapia (Oreochromis aureus: Steindachner 1864), in Lake Edku, Egypt. Global Veterinaria 3: 91–97.
Bartley, D. N., K. Rana & A. J. Immink, 2001. The use of inter-specific hybrids in aquaculture and fisheries. Reviews in Fish Biology and Fisheries 10: 325–337.
Bolnick, D. I. & T. J. Near, 2005. Tempo of hybrid inviability in centrarchid fishes (Teleostei: Centrarchidae). Evolution 59: 1754–1767.
Canonico, C. G., A. Arthington, J. K. McCrary & M. L. Thieme, 2005. The effect of introduced tilapias on native biodiversity. Aquatic Conservation: Marine and Freshwater Ecosystems 15: 463–483.
Clavero, P. M. & E. Garcia-Bethou, 2005. Invasive species are a leading cause of animal extinctions. Trends in Ecology and Evolution 20: 110.
Cox, J. G. & S. I. Lima, 2006. Naïveté and an aquatic-terrestrial dichotomy in the effects of introduced predators. Trends in Ecology and Evolution 21: 676–680.
Crooks, J. A. & M. E. Soulé, 1999. Lag times in population explosions of invasive species: causes and implications. In Sandlund, O. T., P. J. Schei & A. Viken (eds), Invasive Species and Biodiversity Management. Kluwer Academic Publishers, The Netherlands: 103–125.
D’Amato, M. E., M. M. Esterhuyse, B. C. W. van der Waal, D. Brink & F. A. M. Volckaert, 2007. Hybridization and phylogeography of the Mozambique tilapia Oreochromis mossambicus in southern Africa evidenced by mitochondrial and microsatellite DNA genotyping. Conservation Genetics 8: 475–488.
Deines, A. M., I. Bbole, C. Katongo, J. L. Feder & D. M. Lodge, 2014. Hybridisation between non-indigenous Oreochromis niloticus in the Kafue river, Zambia. African Journal of Aquatic Science 39: 23–34.
Deines, A. M., M. E. Wittmann, J. M. Deines & D. M. Lodge, 2016. Tradeoffs among ecosystem services associated with global tilapia introductions. Reviews in Fisheries Science and Aquaculture 24: 178–191.
Doupé, R. G., J. Schaffer, M. J. Knott & D. W. Burrows, 2009. How might an exotic fish disrupt spawning success in a sympatric native species? Marine and Freshwater Research 60: 379–383.
Drake, J. M. & D. M. Lodge, 2003. Global hotspots of biological invasion: evaluating options for ballast-water management. Proceedings of the Royal Society London, Biology 271: 575–580.
Excoffier, L. & H. E. L. Lische, 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567.
FAO, 2012. The state of world fisheries and aquaculture 2012. Rome 200. http://www.fao.org/docrep/016/i2727e/i2727e00.htm
Genner, M. J., P. Nichols, G. R. Carvalho, R. L. Robinson, P. W. Shaw & G. F. Turner, 2007. Reproductive isolation among deep-water cichlid fishes of Lake Malawi differing in monochromatic male breeding dress. Molecular Ecology 16: 651–662.
Genner, M. J., E. Connell, A. Shechonge, A. Smith, J. Swanstrom, S. Mzighani, A. Mwijage, B. P. Ngatunga & G. F. Turner, 2013. Nile tilapia invades the Lake Malawi catchment. African Journal of Aquatic Science 38: 85–90.
Gregg, R. E., J. H. Howard & F. Shonhiwa, 1998. Introgressive hybridization of tilapia in Zimbabwe. Journal of Fish Biology 52: 1–10.
Hall, R. J., 2016. Hybridization helps colonizers become conquerors. Proceedings of the National Academy of Sciences USA 113: 9963–9964.
Hasselman, D. J., E. E. Argo, M. C. McBride, P. Bentzen, T. F. Schultz, A. A. Perez-Umphrey & E. P. Palkovacs, 2014. Human disturbance causes the formation of a hybrid swarm between two naturally sympatric fish species. Molecular Ecology 23: 1137–1152.
Hayden, B., D. Pulcini, M. Kelly-Quinn, M. O’Grady, J. Caffrey, A. McGrath & S. Mariani, 2010. Hybridisation between two cyprinid fishes in a novel habitat: genetics, morphology and life-history traits. BMC Evolutionary Biology 10: 169.
Heck, S., C. Béné & R. Reyes-Gaskin, 2007. Investing in African fisheries: building links to the millennium development goals. Fish and Fisheries 8: 211–226.
Hill, J. E. & C. E. Cichra, 2005. Eradication of reproducing populations of convict cichlids, Cichlasoma nigrofasciatum (Cichlidae), in North-Central Florida. Florida Scientist 68: 65–74.
Hill, J. E. & J. Sowards, 2015. Successful eradication of the non-native loricaridd catfish Pterygolichthys disjunctivus from the Rainbow River, Florida. Management of Biological Invasions 6: 311–317.
Hulme, P. E., S. Bacher, M. Kenis, S. Klotz, I. Kühn, D. Minchin, W. Nentwig, S. Olenin, V. Panov, J. Pergl & P. Pyšek, 2008. Grasping at the routes of biological invasions: a framework for integrating pathways into policy. Journal of Applied Ecology 45: 403–414.
Jombart, T. & I. Ahmed, 2011. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27: 3070–3071.
Kamal, A. H. M. M. & G. C. Mair, 2005. Salinity tolerance in superior genotypes of tilapia, Oreochromis niloticus, Oreochromis mossambicus and their hybrids. Aquaculture 247: 189–201.
Katunzi, E. F. B. & M. A. Kishe, 2004. Changes in population structures of the major species in selected satellite lakes around Lake Victoria following changes in fishing effort. Tanzania Journal of Science 30: 53–64.
Klingenberg, C. P., 2011. MorphoJ: an integrated software package for geometric morphometrics. Molecular Ecology Resources 11: 353–357.
Kolding, J., M. Medard, O. Mkumbo & P. A. M. van Zwieten, 2014. Status, trends and management of the Lake Victoria fisheries. In Inland fisheries evolution and management. Case studies from four continents. FAO: 49–62.
Kopelman, N. M., J. Mayzel, M. Jakobsson, N. A. Rosenberg & I. Mayrose, 2015. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Molecular Ecology Resources 15: 1179–1191.
Leprieur, F., O. Beauchard, S. Blanchet, T. Oberdorff & S. Brosse, 2008. Fish invasions in the world’s river systems, when natural process are blurred by human activities. PloS Biology 6: e28.
Leprieur, F., S. Brosse, E. Garcia-Berthou, T. Oberdorff, J. D. Olden & C. R. Townsend, 2009. Scientific uncertainty and the assessment of risks posed by non-native freshwater fishes. Fish and Fisheries 10: 88–97.
Levin, D. A., J. Francisco-Ortega & R. K. Jansen, 1996. Hybridisation and the extinction of rare plant species. Conservation Biology 10: 10–16.
Levine, J. M. & C. M. D’Antonio, 2003. Forecasting biological invasions with increasing international trade. Conservation Biology 17: 322–326.
Lind, C. E., R. E. Brummett & R. W. Ponzoni, 2012. Exploitation and conservation of fish genetic resources in Africa: issues and priorities for aquaculture development and research. Reviews in Aquaculture 4: 125–141.
Lowe, S., M. Browne, S. Boudjelas & M. De Poorter, 2000. 100 of the world’s worst invasive alien species: a selection from the global invasive species database. Invasive Species Specialist Group, Aliens 12: 12.
Lowell, S. J., S. F. Stone & L. Fernandez, 2006. The economic impact of aquatic invasive species: a review of the literature. Agricultural and Resource Economics Review 35: 195–208.
Lowe-McConnell, R., 2006. The Tilapia Trail. MPM Publishing, Ascot.
Maan, M. E., O. Seehausen & T. G. Groothuis, 2017. Differential survival between visual environments supports a role of divergent sensory drive in cichlid fish speciation. American Naturalist 189: 78–85.
McAndrew, B. J., F. R. Roubal, R. J. Roberts, A. M. Bullock & I. M. McEwen, 1988. The genetics and histology of red, blond and associated colour variants in Oreochromis niloticus. Genetics 76: 127–137.
McGee, M. D., J. W. Reustle, C. E. Oufiero & P. C. Wainwright, 2015. Intermediate kinematics produce inferior feeding performance in a classic case of natural hybridization. American Naturalist 186: 807–814.
Meyerson, L. A. & H. A. Mooney, 2007. Invasive alien species in an era of globalisation. Frontiers in Ecology and the Environment 5: 199–208.
Molnar, J. L., R. L. Gamboa, C. Revenga & M. D. Spalding, 2008. Assessing the global threat of invasive species to marine biodiversity. Frontiers in Ecology and the Environment 6: 485–492.
Moralee, R. D., F. H. van der Bank & B. C. W. van der Waal, 2000. Biochemical genetic markers to identify hybrids between the endemic Oreochromis mossambicus and the alien species, O. niloticus (Pisces: Cichlidae). Water SA 26: 263–268.
Muhlfeld, C. C., S. T. Kalinowski, T. E. McMahon, M. L. Taper, S. Painter, R. F. Leary & F. W. Allendorf, 2009. Hybridization rapidly reduces fitness of a native trout in the wild. Biology Letters 5: 328–331.
Muir, J. F., N. Gitongo, I. Omar, V. Poumogne & I. Radwan, 2005. Hidden harvests: unlocking the potential of aquaculture in Africa. Technical Review Paper. NEPAD-Fish for All Summit 22-25.
Musaka, C. G. & F. F. Musonda, 2013. Contribution of small water bodies and small-holder aquaculture towards poverty allevation and enhancing household food security in Zambia. International Journal of Fisheries and Aquaculture 5: 295–302.
Ndiwa, T. C., D. W. Nyingi & J. F. Agnèse, 2014. An important natural genetic resource of Oreochromis niloticus threatened by aquaculture activities in Loboi Drainage, Kenya. PloS ONE 9: e106972.
Nilsson, P. A., K. Hulthén, B. B. Chapman, L. A. Hansson, J. Brodersen, H. Baktoft, J. Vinterstare, C. Brönmark & C. Skov, 2017. Species integrity enhanced by a predation cost to hybrids in the wild. Biology Letters 13: 20170208.
Nyingi, D. W. & J. F. Agnèse, 2007. Recent introgressive hybridisation revealed by exclusive mtDNA transfer from Oreochromis leucostictus to Oreochromis niloticus in Lake Baringo, Kenya. Journal of Fish Biology 70: 148–154.
Perrings, C., K. Dehnen-Schmutz, J. Touza & M. Williamson, 2005. How to manage biological invasions under globalisation. Trends in Ecology and Evolution 20: 212–215.
Pfennig, K. S., A. L. Kelly & A. A. Pierce, 2016. Hybridization as a facilitator of species range expansion. Proceedings of the Royal Society London B 283: 1839.
Pimentel, D., R. Zuniga & D. Morrison, 2005. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecological Economics 52: 273–288.
Pringle, R. M., 2005. The Origins of the Nile Perch in Lake Victoria. BioScience 55: 780–787.
Pritchard, J. K., M. Stephens & P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Rhymer, J. M. & D. Simberloff, 1996. Extinction by hybridisation and introgression. Annual Review of Ecology and Systematics 27: 83–109.
Rognon, X. & R. Guyomard, 2003. Large extent of mitochondrial DNA transfer from Oreochromis aureus to O. niloticus in West Africa. Molecular Ecology 12: 435–445.
Rohlf, F. J., 2005. tpsDig, digitize landmarks and outlines, version 2.05. Department of Ecology and Evolution, State University of New York at Stony Brook.
Romana-Eugia, M. R. R., M. Ikeda, Z. U. Basiao & N. Taniguchi, 2004. Genetic diversity in farmed Asian Nile and red hybrid tilapia stocks evaluated from microsatellite and mitochondrial DNA analysis. Aquaculture 236: 131–150.
Rosenfield, J. A., S. Nolasco, S. Lindauer, C. Sandoval & A. Kodric-Brown, 2004. The role of hybrid vigor in the replacement of Pecos pupfish by its hybrids with sheepshead minnow. Conservation Biology 18: 1589–1598.
Sala, O. E., F. S. Chapin, J. J. Armesto, E. Berlow, J. Bloomfield, R. Dirzo, E. Huber-Sanwald, L. F. Huenneke, R. B. Jackson, A. Kinzig & R. Leemans, 2000. Global biodiversity scenarios for the year 2100. Science 287: 1770–1774.
Scribner, K. T., K. S. Page & M. L. Barton, 2001. Hybridisation in freshwater fishes: a review of case studies and cytonuclear methods of biological inference. Reviews in Fish Biology and Fisheries 10: 293–323.
Seehausen, O., J. J. van Alphen & F. Witte, 1997. Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science 277: 1808–1811.
Seehausen, O., Y. Terai, I. S. Magalhaes, K. L. Carlton, H. D. Mrosso, R. Miyagi, I. van der Slujis, M. V. Schneider, M. E. Maan, H. Tachnida & H. Imai, 2008. Speciation through sensory drive in cichlid fish. Nature 455: 620–626.
Solem, Ø., O. K. Berg, E. Verspoor, K. Hindar, S. O. Karlsson, J. Koksvik, L. Rønning, G. Kjærstad & J. V. Arnekleiv, 2014. Morphological and genetic comparison between naturally produced smolts of Atlantic salmon, brown trout and their hybrids. Fisheries Management and Ecology 21: 357–365.
Stelkens, R. B., C. Schmid, O. Selz & O. Seehausen, 2009. Phenotypic novelty in experimental hybrids is predicted by the genetic distance between species in cichlid fish. BMC Evolutionary Biology 9: 283.
Taylor, B. W. & R. E. Irwin, 2004. Linking economic activities to the distribution of exotic plants. Proceedings of the National Academy of Sciences of the United States of America 101: 17725–17730.
Todesco, M., M. A. Pascual, G. L. Owens, K. L. Ostevik, B. T. Moyers, S. Hübner, S. M. Heredia, M. A. Hahn, C. Caseys, D. G. Bock & L. H. Rieseberg, 2016. Hybridization and extinction. Evolutionary Applications 9: 892–908.
Trewavas, E., 1983. Tilapiine fishes of the genera Sarotherodon, Oreochromis and Danakilia. British Museum (Natural History), London.
Velema, G. J., J. S. Rosenfeld & E. B. Taylor, 2012. Effects of invasive American signal crayfish (Pacifastacus leniusculus) on the reproductive behaviour of threespine stickleback (Gasterosteus aculeatus) sympatric species pairs. Canadian Journal of Zoology 90: 1328–1338.
Westphal, M. I., M. Browne, K. MacKinnon & I. Noble, 2008. The link between international trade and the global distribution of invasive alien species. Biological Invasions 10: 391–398.
Williams, S. L. & E. D. Grosholz, 2008. The invasive species challenge in estuarine and coastal environments: marrying management and science. Estuaries and Coasts 31: 3–20.
Zengeya, T. A., M. P. Robertson, A. J. Booth & C. T. Chimimba, 2013. A qualitative ecological risk assessment of the invasive Nile tilapia, Oreochromis niloticus in a sub-tropical African river system (Limpopo River, South Africa). Aquatic Conservation: Marine and Freshwater Ecosystems 23: 51–64.
Acknowledgements
The work was supported by Royal Society-Leverhulme Trust Africa Awards AA100023 and AA130107 to MJG, BPN and GFT, and a BBSRC award BB/M026736/1 to GFT and MJG. The Tanzania Commission for Science and Technology (COSTECH) provided fieldwork permits, and staff. Tanzania Fisheries Research Institute contributed to fieldwork.
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest editors: S. Koblmüller, R. C. Albertson, M. J. Genner, K. M. Sefc & T. Takahashi / Advances in Cichlid Research III: Behavior, Ecology and Evolutionary Biology
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Bradbeer, S.J., Harrington, J., Watson, H. et al. Limited hybridization between introduced and Critically Endangered indigenous tilapia fishes in northern Tanzania. Hydrobiologia 832, 257–268 (2019). https://doi.org/10.1007/s10750-018-3572-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-018-3572-5