Abstract
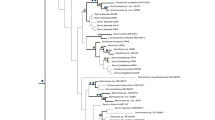
Marine sponges of the class Hexactinellida (glass sponges) are among the most understudied groups of Porifera, and molecular approaches to investigating their evolution have only recently emerged. Although these first results appeared reliable as they largely corroborated morphology-based hypotheses, they were almost exclusively based on ribosomal RNA genes (rDNA) and should, therefore, be further tested with independent types of genetic data, such as protein-coding genes. To this end, we established the mitochondrial-encoded cytochrome oxidase subunit I gene (COI) as an additional marker, and conducted phylogenetic analyses on DNA- and amino-acid level, as well as a supermatrix analysis based on combined COI DNA and rDNA alignments. Furthermore, we increased taxon sampling compared to previous studies by adding seven additional species. The COI-based phylogenies were largely congruent with the rDNA-based phylogeny but suffered from poor bootstrap support for many nodes. However, addition of the COI sequences to the rDNA data set increased resolution of the overall molecular phylogeny. Thus, although obtaining COI sequences from glass sponges turned out to be quite challenging, this gene appears to be a valuable supplement to rDNA data for molecular evolutionary studies of this group. Some implications of our extended phylogeny for the evolution and systematics of Hexactinellida are discussed.




Similar content being viewed by others
References
Abascal, F., R. Zardoya & D. Posada, 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105.
Akaike, H., 1974. A new look at the statistical model identifications. IEEE Transactions on Automatic Control 19: 716–723.
Bebenek, I. G., R. D. Gates, J. Morris, V. Hartenstein & D. K. Jacobs, 2004. sine oculis in basal Metazoa. Development Genes and Evolution 214: 342–351.
Borchiellini, C., N. Boury-Esnault, J. Vacelet & Y. Le Parco, 1998. Phylogenetic analysis of the Hsp70 sequences reveals the monophyly of Metazoa and specific phylogenetic relationships between animals and fungi. Molecular Biology and Evolution 15: 647–655.
Bucklin, A., D. Steinke & L. Blanco-Bercial, 2011. DNA barcoding of marine Metazoa. Annual Review of Marine Science 3: 471–508.
Conejo, M., M. Bertin, S. A. Pomponi & W. R. Ellington, 2008. The early evolution of the phosphagen kinases – insights from choanoflagellate and poriferan arginine kinases. Journal of Molecular Evolution 66: 11–20.
de Queiroz, A. & J. Gatesy, 2007. The supermatrix approach to systematics. Trends in Ecology and Evolution 22: 34–41.
Dohrmann, M., O. Voigt, D. Erpenbeck & G. Wörheide, 2006. Non-monophyly of most supraspecific taxa of calcareous sponges (Porifera, Calcarea) revealed by increased taxon sampling and partitioned Bayesian analysis of ribosomal DNA. Molecular Phylogenetics and Evolution 40: 830–843.
Dohrmann, M., D. Janussen, J. Reitner, A. G. Collins & G. Wörheide, 2008. Phylogeny and evolution of glass sponges (Porifera, Hexactinellida). Systematic Biology 57: 388–405.
Dohrmann, M., A. G. Collins & G. Wörheide, 2009. New insights into the phylogeny of glass sponges (Porifera, Hexactinellida): monophyly of Lyssacinosida and Euplectellinae, and the phylogenetic position of Euretidae. Molecular Phylogenetics and Evolution 52: 257–262.
Dohrmann, M., C. Göcke, D. Janussen, J. Reitner, C. Lüter & G. Wörheide, 2011. Systematics and spicule evolution in dictyonal sponges (Hexactinellida: Sceptrulophora) with description of two new species. Zoological Journal of the Linnean Society (in press).
Erpenbeck, D. & G. Wörheide, 2007. On the molecular phylogeny of sponges (Porifera). Zootaxa 1668: 107–126.
Erpenbeck, D., J. N. A. Hooper & G. Wörheide, 2006. CO1 phylogenies in diploblasts and the ‘Barcoding of Life’ – are we sequencing a suboptimal partition? Molecular Ecology Notes 6: 550–553.
Felsenstein, J., 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791.
Göcke, C. & D. Janussen, 2011. ANT XXIV/2 (SYSTCO) Hexactinellida (Porifera) and bathymetric traits of Antarctic glass sponges (incorporating ANDEEP-material); including an emendation of the rediscovered genus Lonchiphora. Deep-Sea Research Part II: in press.
Gouy, M., S. Guindon & O. Gascuel, 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution 27: 221–224.
Gundacker, D., S. P. Leys, H. C. Schröder, I. M. Müller & W. E. G. Müller, 2001. Isolation and cloning of a C-type lectin from the hexactinellid sponge Aphrocallistes vastus: a putative aggregation factor. Glycobiology 11: 21–29.
Haen, K. M., B. F. Lang, S. A. Pomponi & D. V. Lavrov, 2007. Glass sponges and bilaterian animals share derived mitochondrial genomic features: a common ancestry or parallel evolution? Molecular Biology and Evolution 24: 1518–1527.
Hillis, D. M. & J. J. Bull, 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192.
Huelsenbeck, J. P. & B. Rannala, 2004. Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Systematic Biology 53: 904–913.
Ivanova, N. V., A. V. Borisenko & P. D. N. Hebert, 2009. Express barcodes: racing from specimen to identification. Molecular Ecology Resources 9(Supplement 1): 35–41.
Lanave, C., G. Preparata, C. Saccone & G. Serio, 1984. A new method for calculating evolutionary substitution rates. Journal of Molecular Evolution 20: 86–93.
Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson & D. G. Higgins, 2007. ClustalW and ClustalX version 2.0. Bioinformatics 23: 2947–2948.
Leys, S. P. G. O., G. O. Mackie & H. M. Reiswig, 2007. The biology of glass sponges. Advances in Marine Biology 52: 1–145.
Manuel, M., Y. Le Parco & C. Borchiellini, 2004. Comparative analysis of Brachyury T-domains, with the characterization of two new sponge sequences, from a hexactinellid and a calcisponge. Gene 340: 291–301.
Mehl, D., 1992. Die Entwicklung der Hexactinellida seit dem Mesozoikum. Paläobiologie, Phylogenie und Evolutionsökologie. Berliner Geowissenschaftliche Abhandlungen Reihe E: Paläobiologie 2: 1–164.
Philippe, H., R. Derelle, P. Lopez, K. Pick, C. Borchiellini, N. Boury-Esnault, J. Vacelet, E. Renard, E. Houliston, E. Quéinnec, C. Da Silva, P. Wincker, H. Le Guyader, S. Leys, D. J. Jackson, F. Schreiber, D. Erpenbeck, B. Morgenstern, G. Wörheide & M. Manuel, 2009. Phylogenomics revives traditional views on deep animal relationships. Current Biology 19: 706–712.
Pick, K. S., H. Philippe, F. Schreiber, D. Erpenbeck, D. J. Jackson, P. Wrede, M. Wiens, A. Alié, B. Morgenstern, M. Manuel & G. Wörheide, 2010. Improved phylogenomic taxon sampling noticeably affects non-bilaterian relationships. Molecular Biology and Evolution 27: 1983–1987.
Reiswig, H. M., 2002. Family Tretodictyidae Schulze, 1886. In Hooper, J. N. A. & R. W. M. van Soest (eds), Systema Porifera. A Guide to the Classification of Sponges. Plenum, New York: 1341–1354.
Rokas, A., N. King, J. Finnerty & S. B. Carroll, 2003. Conflicting phylogenetic signals at the base of the metazoan tree. Evolution and Development 5: 346–359.
Rosengarten, R. D., E. A. Sperling, M. A. Moreno, S. P. Leys & S. L. Dellaporta, 2008. The mitochondrial genome of the hexactinellid sponge Aphrocallistes vastus: evidence for programmed translational frameshifting. BMC Genomics 9: 33.
Savill, N. J., D. C. Hoyle & P. G. Higgs, 2001. RNA sequence evolution with secondary structure constraints: comparison of substitution rate models using maximum-likelihood methods. Genetics 157: 399–411.
Siddall, M. E., F. M. Fontanella, S. C. Watson, S. Kvist & C. Erséus, 2009. Barcoding bamboozled by bacteria: convergence to metazoan mitochondrial primer targets by marine microbes. Systematic Biology 58: 445–451.
Sperling, E. A., K. J. Peterson & D. Pisani, 2009. Phylogenetic-signal dissection of nuclear housekeeping genes supports the paraphyly of sponges and the monophyly of Eumetazoa. Molecular Biology and Evolution 26: 2261–2274.
Stamatakis, A., 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690.
Stamatakis, A., P. Hoover & J. Rougemont, 2008. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57: 758–771.
Tabachnick, K. R., 2002a. Lyssacinosida incertae sedis. In Hooper, J. N. A. & R. W. M. van Soest (eds), Systema Porifera. A Guide to the Classification of Sponges. Plenum, New York: 1506–1509.
Tabachnick, K. R., 2002b. Family Rossellidae Schulze, 1885. In Hooper, J. N. A. & R. W. M. van Soest (eds), Systema Porifera. A Guide to the Classification of Sponges. Plenum, New York: 1441–1505.
Tabachnick, K. R., 2002c. Family Euplectellidae Gray, 1867. In Hooper, J. N. A. & R. W. M. van Soest (eds), Systema Porifera. A Guide to the Classification of Sponges. Plenum, New York: 1388–1434.
Tabachnick, K. R. & H. M. Reiswig, 2002. Dictionary of Hexactinellida. In Hooper, J. N. A. & R. W. M. van Soest (eds), Systema Porifera. A Guide to the Classification of Sponges. Plenum, New York: 1224–1229.
Yang, Z., 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. Journal of Molecular Evolution 39: 306–314.
Acknowledgments
Work at the Wörheide lab was supported by the Deutsche Forschungsgemeinschaft (DFG; project Wo896/5-3). The DFG is further acknowledged for funding the Deep Down Under expedition (Project Wo896/7-1; www.deepdownunder.de). During writing of this manuscript, MD was supported by a Postdoctoral Fellowship of the Smithsonian Institution. Work at the Lavrov lab was supported by the National Science Foundation (NSF) through the Poriferan Tree of Life project (DEB-0828783; https://www.portol.org/). Shirley Pomponi (HBOI), Sally Leys, and Jean Vacelet are acknowledged for providing additional specimens. Our warmest thanks go to the organizers of the 8th World Sponge Conference for making this great meeting possible. Allen Collins and three anonymous reviewers provided comments that led to improvement of this manuscript. Finally, we would like to thank Alexandros Stamatakis for implementing paired-sites models in his amazing program, RAxML.
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest editors: M. Maldonado, X. Turon, M. A. Becerro & M. J. Uriz / Ancient animals, new challenges: developments in sponge research
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dohrmann, M., Haen, K.M., Lavrov, D.V. et al. Molecular phylogeny of glass sponges (Porifera, Hexactinellida): increased taxon sampling and inclusion of the mitochondrial protein-coding gene, cytochrome oxidase subunit I. Hydrobiologia 687, 11–20 (2012). https://doi.org/10.1007/s10750-011-0727-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-011-0727-z