Abstract
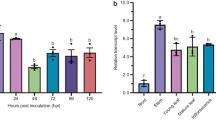
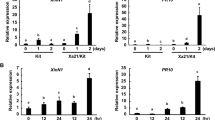
SNAREs play important roles in plant responses to various biotic and abiotic stresses. OsSNAP32 is a SNAP25-type SNARE protein-encoding gene isolated from rice. OsSNAP32 is ubiquitously expressed in the various tissues of blast-resistant rice landrace Heikezijing and blast-susceptible Suyunuo, with higher expression in Heikezijing and induced expression in rice seedlings inoculated by the blast pathogen (Magnaporthe oryzae) race Hoku1. OsSNAP32-overexpressing transgenic lines in Suyunuo increased resistance to blast, with fewer lesions in the inoculated leaves compared with the non-transgenic control Suyunuo. OsSNAP32 RNAi transgenic lines in Heikezijing decreased resistance to blast, with some typical lesions in the inoculated leaves compared with the non-transgenic control Heikezijing. These results suggested that OsSNAP32 might be involved in rice resistance to blast.






Similar content being viewed by others
Abbreviations
- Dpi:
-
Days past inoculation
- Hpi:
-
Hours past inoculation
- SD:
-
Standard deviation
- cDNA:
-
Complementary DNA
- CTAB:
-
Hexadecyl trimethyl ammonium bromide
References
Abrahams BS et al (2003) Metaphase fishing of transgenic mice recommended: fish and sky define BAC-mediated balanced translocation. Genesis (New York, NY: 2000) 36:134–141
Ahmed I, Islam M, Arshad W, Mannan A, Ahmad W, Mirza B (2009) High-quality plant DNA extraction for PCR: an easy approach. J Appl Genet 50:105–107
Bao YM, Wang JF, Huang J, Zhang HS (2008) Molecular cloning and characterization of a novel SNAP25-type protein gene OsSNAP32 in rice (Oryza sativa L.). Mol Biol Rep 35:145–152
Blatt MR (2002) Toward understanding vesicle traffic and the guard cell model. New Phytol 153:405–413
Chandler KJ, Chandler RL, Broeckelmann EM, Hou Y, Southard-Smith EM, Mortlock DP (2007) Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm Genome 18:693–708
Chen YA, Scales SJ, Patel SM, Doung YC, Scheller RH (1999) SNARE complex formation is triggered by Ca2+ and drives membrane fusion. Cell 97:165–174
Chiang C et al (2012) Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet 44:390–397
Collins NC et al (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425:973–977
Fasshauer D, Sutton RB, Brunger AT, Jahn R (1998) Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA 95:15781–15786
Fukuda R et al (2000) Functional architecture of an intracellular membrane t-SNARE. Nature 407:198–202
Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE (1997) Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90:523–535
Heese M, Gansel X, Sticher L, Wick P, Grebe M, Granier F, Jurgens G (2001) Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J Cell Biol 155:239–249
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J: Cell Mol Biol 6:271–282
Jahn R, Scheller RH (2006) SNAREs–engines for membrane fusion. Nat Rev Mol Cell Biol 7:631–643
Jahn R, Sudhof TC (1999) Membrane fusion and exocytosis. Annu Rev Biochem 68:863–911
Kasai F, Yoshihara M, Matsukuma S, O’Brien P, Ferguson-Smith MA (2007) Emergence of complex rearrangements at translocation breakpoints in a transgenic mouse; implications for mechanisms involved in the formation of chromosome rearrangements. Cytogenet Genome Res 119:83–90
Kato T, Morita MT, Fukaki H, Yamauchi Y, Uehara M, Niihama M, Tasaka M (2002) SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14:33–46
Kwon C et al (2008) Co-option of a default secretory pathway for plant immune responses. Nature 451:835–840
Ladunga I (2007) More complete gene silencing by fewer siRNAs: transparent optimized design and biophysical signature. Nucleic Acids Res 35:433–440
Le Saux A, Houdebine LM, Jolivet G (2010) Chromosome integration of BAC (bacterial artificial chromosome): evidence of multiple rearrangements. Transgenic Res 19:923–931
Leyman B, Geelen D, Quintero FJ, Blatt MR (1999) A tobacco syntaxin with a role in hormonal control of guard cell ion channels. Science 283:537–540
Li PF, Zhai HQ, Zhang HS, Lu ZQ, Chen ZY, Wang FM (1999) Inheritance of blast resistance in two Japonica rice landraces from Taihu lake area. Chin J Rice Sci 13:11–14
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408
Lu C, Shen L, Tan Z, Xu Y, He P, Chen Y, Zhu L (1996) Comparative mapping of QTLs for agronomic traits of rice across environments using a doubled haploid population. TAG Theor Appl Genet Theoretische und angewandte Genetik 93:1211–1217
Lukowitz W, Mayer U, Jurgens G (1996) Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84:61–71
Ma K, Xiao J, Li X, Zhang Q, Lian X (2009) Sequence and expression analysis of the C3HC4-type RING finger gene family in rice. Gene 444:33–45
Mackill DJ, Bonman JM (1992) Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 82:746–749
Nam KH, Cho A, Kwon JY, Park YW, Kim YH (2012) Feasibility of measuring 3-dimensional renal parenchymal volume to predict postnatal renal function in near-term fetuses with congenital hydronephrosis: a preliminary study. J Ultrasound Med 31:955–962
Poirier MA, Xiao W, Macosko JC, Chan C, Shin YK, Bennett MK (1998) The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat Struct Biol 5:765–769
Pratelli R, Sutter JU, Blatt MR (2004) A new catch in the SNARE. Trends Plant Sci 9:187–195
Shi XL, Wang JF, Bao YM, Li PF, Xie LJ, Huang J, Zhang HS (2010) Identification of the quantitative trait loci in Japonica rice landrace Heikezijing responsible for broad-spectrum resistance to rice blast. Phytopathology 100:822–829
Sutter JU, Campanoni P, Blatt MR, Paneque M (2006) Setting SNAREs in a different wood. Traffic (Copenhagen, Denmark) 7:627–638
Sutton RB, Fasshauer D, Jahn R, Brunger AT (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 angstrom resolution. Nature 395:347–353
Suzuki M, Kondo S, Pei Z, Maekawa A, Saito I, Kanegae Y (2015) Preferable sites and orientations of transgene inserted in the adenovirus vector genome: the E3 site may be unfavorable for transgene position. Gene Ther 22:421–429
Vert JP, Foveau N, Lajaunie C, Vandenbrouck Y (2006) An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinform 7:520
Wang M, Chen C, Xu YY, Jiang RX, Han Y, Xu ZH, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22:409–417
Wick P, Gansel X, Oulevey C, Page V, Studer I, Durst M, Sticher L (2003) The expression of the t-SNARE AtSNAP33 is induced by pathogens and mechanical stimulation. Plant Physiol 132:343–351
Yano D, Sato M, Saito C, Sato MH, Morita MT, Tasaka M (2003) A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc Natl Acad Sci USA 100:8589–8594
Zhu J et al (2002) OSM1/SYP61: a syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell 14:3009–3028
Acknowledgments
This research has been supported by Grants the National Key Project for Transgenic Crops (2014ZX08009-001B, 2014ZX08009-003-001-010), the Natural Science Foundation of China (30900888 and 31171516), the Fundamental Research Funds for the Central Universities (KYZ201302), Jiangsu Agriculture science and technology innovation fund (CX(12)1003-3, CX(15)1054). We wish to thank Prof. Zhongzhuan Lin, CAAS, China for providing the Japanese blast race Hoku1 and Prof. Zhiyi Chen, Prof. Yongfeng Liu, JAAS, China for providing six Chinese blast races, i.e., 191ZB13, 97-2ZC15, 35-1ZD1, 55-1ZE3, 42-2ZF1 and 113ZG1.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Luo, J., Zhang, H., He, W. et al. OsSNAP32, a SNAP25-type SNARE protein-encoding gene from rice, enhanced resistance to blast fungus. Plant Growth Regul 80, 37–45 (2016). https://doi.org/10.1007/s10725-016-0152-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-016-0152-4