Abstract
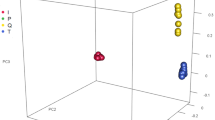
Up to date, the scarcity of publicly available complete mitochondrial sequences for European wild pigs hampers deeper understanding about the genetic changes following domestication. Here, we have assembled 26 de novo mtDNA sequences of European wild boars from next generation sequencing (NGS) data and downloaded 174 complete mtDNA sequences to assess the genetic relationship, nucleotide diversity, and selection. The Bayesian consensus tree reveals the clear divergence between the European and Asian clade and a very small portion (10 out of 200 samples) of maternal introgression. The overall nucleotides diversities of the mtDNA sequences have been reduced following domestication. Interestingly, the selection efficiencies in both European and Asian domestic pigs are reduced, probably caused by changes in both selection constraints and maternal population size following domestication. This study suggests that de novo assembled mitogenomes can be a great boon to uncover the genetic turnover following domestication. Further investigation is warranted to include more samples from the ever-increasing amounts of NGS data to help us to better understand the process of domestication.






Similar content being viewed by others
References
Amaral AJ, Ferretti L, Megens HJ, Crooijmans RP, Nie H, Ramos-Onsins SE, Perez-Enciso M, Schook LB, Groenen MA (2011) Genome-wide footprints of pig domestication and selection revealed through massive parallel sequencing of pooled DNA. PLoS One 6:e14782
Björnerfeldt S, Webster MT, Vilà C (2006) Relaxation of selective constraint on dog mitochondrial DNA following domestication. Genome Res 16:990–994
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Darwin CR (1868) Variation of plants and animals under domestication
Frantz LA, Schraiber JG, Madsen O, Megens H-J, Cagan A, Bosse M, Paudel Y, Crooijmans RP, Larson G, Groenen MA (2015) Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat Genet 47:1141–1148
Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, Rogel-Gaillard C, Park C, Milan D, Megens H-J (2012) Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491:393–398
Isaac E (1974) History of domestic mammals in Central and Eastern Europe by S. Bökönyi. Euromoney 144:94–98
Jin L, Zhang M, Ma J, Zhang J, Zhou C, Liu Y, Wang T, Jiang AA, Chen L, Wang J (2012) Mitochondrial DNA evidence indicates the local origin of domestic pigs in the upstream region of the Yangtze River. PLoS One 7:e51649
Kimura M (1962) On the probability of fixation of mutant genes in a population. Genetics 47:713
Kimura M, 1984. The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870
Kusza S, Podgórski T, Scandura M, Borowik T, Jávor A, Sidorovich VE, Bunevich AN, Kolesnikov M, Jędrzejewska B (2014) Contemporary genetic structure, phylogeography and past demographic processes of wild boar Sus scrofa population in Central and Eastern Europe. PLoS One 9:e91401
Larson G, Dobney K, Albarella U, Fang M, Matisoo-Smith E, Robins J, Lowden S, Finlayson H, Brand T, Willerslev E (2005) Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 307:1618–1621
Larson G, Albarella U, Dobney K, Rowleyconwy P, Schibler J, Tresset A, Vigne JD, Edwards CJ, Schlumbaum A, Dinu A (2007) Ancient DNA, pig domestication, and the spread of the Neolithic into Europe. Proc Natl Acad Sci USA 104:15276
Larson G, Liu R, Zhao X, Yuan J, Fuller D, Barton L, Dobney K, Fan Q, Gu Z, Liu X-H (2010) Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proc Natl Acad Sci 107:7686–7691
Laval G, Iannuccelli N, Legault C, Milan D, Groenen MA, Giuffra E, Andersson L, Nissen PH, Jørgensen CB, Beeckmann P (2000) Genetic diversity of eleven European pig breeds. Genet Select Evol 32:187–203
Li M, Jin L, Ma J, Tian S, Li R, Li X (2016) Detecting mitochondrial signatures of selection in wild Tibetan pigs and domesticated pigs. Mitochondrial DNA 27:747
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lin CS, Sun YL, Liu CY, Yang PC, Chang LC, Cheng IC, Mao SJT, Huang MC (1999) Complete nucleotide sequence of pig (Sus scrofa) mitochondrial genome and dating evolutionary divergence within Artiodactyla. Gene 236:107–114
McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32:W20–W25
Mitterboeck TF, Adamowicz SJ (2013) Flight loss linked to faster molecular evolution in insects. Proc R Soc Lond B Biol Sci 280:20131128
Panoraia A, Alexer T, Spiros P, Evangelos C, Petros P, Nikolaos P, Greger L, Abatzopoulos TJ, Costas T (2012) The Balkans and the colonization of Europe: the post-glacial range expansion of the wild boar, Sus scrofa. J Biogeogr 39:713–723
Petit N, Barbadilla A (2009) Selection efficiency and effective population size in Drosophila species. J Evol Biol 22:515–526
Ratnakumar A (2013) Detecting signatures of selection within the dog genome. Acta Universitatis Upsaliensis
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Shen Y-Y, Shi P, Sun Y-B, Zhang Y-P (2009) Relaxation of selective constraints on avian mitochondrial DNA following the degeneration of flight ability. Genome Res 19:1760–1765
Stewart JB, Freyer C, Elson JL, Larsson N-G (2008) Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat Rev Genet 9:657–662
Tanaka K, Iwaki Y, Takizawa T, Dorji T, Tshering G, Kurosawa Y, Maeda Y, Mannen H, Nomura K, Dang VB (2010) Mitochondrial diversity of native pigs in the mainland South and South-east Asian countries and its relationships between local wild boars. Anim Sci J 79:417–434
Ursing BM, Arnason U (1998) The complete mitochondrial DNA sequence of the pig (Sus scrofa). J Mol Evol 47:302–306
Vilaça ST, Biosa D, Zachos F, Iacolina L, Kirschning J, Alves PC, Paule L, Gortazar C, Mamuris Z, Jędrzejewska B (2014) Mitochondrial phylogeography of the European wild boar: the effect of climate on genetic diversity and spatial lineage sorting across Europe. J Biogeogr 41:987–998
Wang Z, Yonezawa T, Liu B, Ma T, Shen X, Su J, Guo S, Hasegawa M, Liu J (2011) Domestication relaxed selective constraints on the yak mitochondrial genome. Mol Biol Evol 28:1553–1556
Wang G-D, Xie H-B, Peng M-S, Irwin D, Zhang Y-P (2014) Domestication genomics: evidence from animals. Annu Rev Anim Biosci 2:65–84
Wangsheng X (2004) Introduction and domestication of European breeds of pig in modern China. Anc Mod Agric 1:008
Wu G-S, Yao Y-G, Qu K-X, Ding Z-L, Li H, Palanichamy MG, Duan Z-Y, Li N, Chen Y-S, Zhang Y-P (2007) Population phylogenomic analysis of mitochondrial DNA in wild boars and domestic pigs revealed multiple domestication events in East Asia. Genome Biol 8:1
Xu L, Bickhart DM, Cole JB, Schroeder SG, Song J, Tassell CP, Sonstegard TS, Liu GE (2015) Genomic signatures reveal new evidences for selection of important traits in domestic cattle. Mol Biol Evol 32:711
Yang Z (2006) Computational molecular evolution. Oxford University Press, Oxford
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang S-L, Wang Z-G, Liu B, Zhang G-X, Zhao S-H, Yu M, Fan B, Li M-H, Xiong T-A, Li K (2003) Genetic variation and relationships of eighteen Chinese indigenous pig breeds. Genet Select Evol 35:657
Acknowledgements
This Project was supported by National Natural Science Foundation of China (NSFC-CGIAR, No. 31361140365), Foshan University Initiative Scientific Research Program (No. 31702088) and the Fundamental Research Funds for the Central Universities (Grant ID: 2662015JC010 and 2662017JC027).
Author information
Authors and Affiliations
Contributions
KL and SZ conceived and designed the experiments; PN, CZ and HY downloaded and compiled the public data; PN, JHC, AAB, JL, XL and XD analyzed the data; AAB, PN and JHC wrote the manuscript; KL, and SZ finalized the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest and responsible for the content and authorship of this paper.
Rights and permissions
About this article
Cite this article
Ni, P., Bhuiyan, A.A., Chen, JH. et al. De novo assembly of mitochondrial genomes provides insights into genetic diversity and molecular evolution in wild boars and domestic pigs. Genetica 146, 277–285 (2018). https://doi.org/10.1007/s10709-018-0018-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-018-0018-y