Abstract
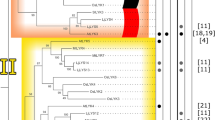
Lipases are physiologically important and ubiquitous enzymes that share a conserved domain and are classified into eight different families based on their amino acid sequences and fundamental biological properties. The Lipase3 family of lipases was reported to possess a canonical fold typical of α/β hydrolases and a typical catalytic triad, suggesting a distinct evolutionary origin for this family. Genes in the Lipase3 family do not have the same functions, but maintain the conserved Lipase3 domain. There have been extensive studies of Lipase3 structures and functions, but little is known about their evolutionary histories. In this study, all lipases within five plant species were identified, and their phylogenetic relationships and genetic properties were analyzed and used to group them into distinct evolutionary families. Each identified lipase family contained at least one dicot and monocot Lipase3 protein, indicating that the gene family was established before the split of dicots and monocots. Similar intron/exon numbers and predicted protein sequence lengths were found within individual groups. Twenty-four tandem Lipase3 gene duplications were identified, implying that the distinctive function of Lipase3 genes appears to be a consequence of translocation and neofunctionalization after gene duplication. The functional genes EDS1, PAD4, and SAG101 that are reportedly involved in pathogen response were all located in the same group. The nucleotide diversity (Dxy) and the ratio of nonsynonymous to synonymous nucleotide substitutions rates (Ka/Ks) of the three genes were significantly greater than the average across the genomes. We further observed evidence for selection maintaining diversity on three genes in the Toll-Interleukin-1 receptor type of nucleotide binding/leucine-rich repeat immune receptor (TIR-NBS LRR) immunity-response signaling pathway, indicating that they could be vulnerable to pathogen effectors.

Similar content being viewed by others
References
Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 95:10306
Arpigny JL, Jaeger KE (1999) Bacterial lipolytic enzymes: classification and properties. Biochem J 343(Pt 1):177
Caldwell KS, Michelmore RW (2009) Arabidopsis thaliana genes encoding defense signaling and recognition proteins exhibit contrasting evolutionary dynamics. Genetics 181:671–684
Falk A, Feys BJ, Frost LN, Jones JD, Daniels MJ, Parker JE (1999) EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA 96:3292
Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. Embo J 20:5400–5411
Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C et al (2005) Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17:2601
He Y, Gan S (2002) A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 14:805–815
Heyndrickx KS, Vandepoele K (2012) Systematic Identification of functional plant modules through the integration of complementary data sources. Plant Physiol 159:884–901
Hyun Y, Choi S, Hwang HJ, Yu J et al (2008) Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Dev Cell 14:183–192
Ishiguro S, Kawaioda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13:2191–2209
Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE et al. (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci 96:13583
Masomian M, Rahman RNZRA., Salleh AB, Basri M (2016) Analysis of comparative sequence and genomic data to verify phylogenetic relationship and explore a new subfamily of bacterial lipases. Plos One 11:e0149851
Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM et al (1992) The alpha/beta hydrolase fold. Protein Eng 5:197
Schiml S, Fauser F, Puchta H (2016) Repair of adjacent single-strand breaks is often accompanied by the formation of tandem sequence duplications in plant genomes. Proc Natl Acad Sci 113:7266
Schrag JD, Cygler M (1997) Lipases and alpha/beta hydrolase fold. Methods Enzymol 284:85
Shah J (2005) Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Annu Rev Phytopathol 43:229–260
Sugihara A, Hata S, Shimada Y, Goto K, Tsunasawa S, Tominaga Y (1993) Characterization of Geotrichum candidum lipase III with some preference for the inside ester bond of triglyceride. Appl Microbiol Biotechnol 40:279
Szalontai B, Stranczinger S, Palfalvi G, Mauch-Mani B, Jakab G (2012) The taxon-specific paralogs of grapevine PRLIP genes are highly induced upon powdery mildew infection. J Plant Physiol 169:1767–1775
Van dPY, Fawcett JA, Proost S, Sterck L, Vandepoele, K (2009) The flowering world: a tale of duplications. Trends Plant Sci 14:680
Wiermer M, Feys BJ, Parker JE (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8:383–389
Wong H, Schotz MC (2002) The lipase gene family. J Lipid Res 43:993
Funding
This work was funded by Jiangsu Postdoctoral Science Foundation funded project (1601080C), Jiangsu University Natural Science Foundation funded project (17KJB310015), and Research Foundation for Talented Scholars in Xuzhou Medical University (D2015001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors indicate no potential conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, D., Zhang, L., Hu, J. et al. Comparative genomic analysis of the Lipase3 gene family in five plant species reveals distinct evolutionary origins. Genetica 146, 179–185 (2018). https://doi.org/10.1007/s10709-018-0010-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-018-0010-6