Abstract
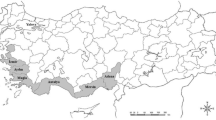
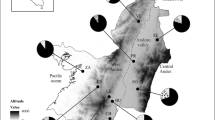
The study examined the genetic diversity and demographic history of Bactrocera dorsalis, a destructive and polyphagous insect pest of fruit crops in diverse geographic regions of India. 19 widely dispersed populations of the fly from India and other Asian countries were analysed using partial sequences of mitochondrial cytochrome oxidase I (cox1) and NADH dehydrogenase 1 (nad1) genes to investigate genetic diversity, genetic structure, and demographic history in the region. Genetic diversity indices [number of haplotypes (H), haloptype diversity (Hd), nucleotide diversity (π) and average number of nucleotide difference (k)] of populations revealed that B. dorsalis maintains fairly high level of genetic diversity without isolation by distance among the geographic regions. Demographic analysis showed significant (negative) Tajimas’ D and Fu’s F S with non significant sum of squared deviations (SSD) values, which indicate the possibility of recent sudden expansion of species and is further supported through distinctively star-like distribution structure of haplotypes among populations. Thus, the results indicate that both ongoing and historical factors have played important role in determining the genetic structure and diversity of the species in India. Consequently, sterile insect technique (SIT) could be a possible management strategy of species in the regions.



Similar content being viewed by others
References
Aketarawong N, Bonizzoni M, Thanaphum S, Gomulski LM, Gasperi G, Malacrida AR, Gugliemino CR (2007) Inferences on the population structure and colonization process of the invasive oriental fruit fly, Bactrocera dorsalis (Hendel). Mol Ecol 16(17):3522–3532
Aketarawong N, Guglielmino CR, Karam N, Falchetto M, Manni M, Scolari F, Gomulski LM, Gasperi G, Malacrida AR (2014) The oriental fruit fly Bactrocera dorsalis s.s. in East Asia; disentangling the different force promoting the invasion and shaping the genetic make-up of populations. Genetica 142:201–213
Bandelt H, Forster P, Roehl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16(1):37–48
Bezzi M (1913) Indian Tephritids (fruit flies) in the collection of the Indian Museum, Calcutta. Mem Indian Mus 3:153–175
Bezzi M (1916) On the fruit flies of the genus Dacus (s.l.) occurring in India, Burma, and Ceylon. Bull Entomol Res 7:99–121
Blackburn TM, Pysek P, Bacher S, Carlton JT, Duncan RP, Jarosik V, Wilson JR, Richardson DM (2011) A proposed unified framework for biological invasions. Trends Ecol Evol 26:333–339
Bryja J, Smith C, Konecny A, Reichaid M (2010) Range-wide population genetic structure of the European bitterling (Rhodeus amarus) based on microsatellite and mitochondrial DNA analysis. Mol Ecol 19:4708–4722
CABI (2016) Bactrocera dorsalis (oriental fruit fly): invasive species compendium datasheets, maps, images, abstracts and full text on invasive species of the world. http://www.cabi.org/isc/datasheet/17685. Accessed 1 Aug 2016
Chen CC, Dong YJ, Li CT, Liu KY (2006) Movement of the oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae), in a guava orchard with special reference to its population changes. Formos Entomol 26:143–159
Choudhary JS, Kumari A, Das B, Maurya S, Kumar S (2012) Diversity and population dynamic of fruit flies species in methyl eugenol based parapheromone traps in Jharkhand region of India. The Ecoscan 1:57–60
Choudhary JS, Naaz N, Prabhakar CS, Das B, Maurya S, Kumar S (2014) Field guide for identification of Fruit fly species of Genus Bactrocera prevalent in and around mango orchards. Technical booklets No. R-43/Ranchi-16. ICAR Research Complex for Eastern Region, Research Centre, Ranchi, pp 1–16
Drew RAI, Raghu S (2002) The fruit fly fauna (Diptera: Tephritidae: Dacinae) of the rainforest habitat of the Western Ghats, India. Raffles Bull Zool 50(2):327–352
Estoup A, Guillemaud T (2010) Reconstructing routes of invasion using genetic data: why, how and so what? Mol Ecol 19:4113–4130
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5 a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3):564–567
Finn DS, Theobald DM, Black IV, Poff NL (2006) Spatial population genetic structure and limited dispersal in a Rocky Mountain alpine stream insect. Mol Ecol 15:3553–3566
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Grant WS, Bowen BW (1998) Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J Hered 89:415–426
Grapputo A, Bisazza A, Pilastro A (2006) Invasion success despite reduction of genetic diversity in the European populations of eastern mosquito fish (Gambusia holbrooki). Ital J Zool 73:67–73
Hardy DE (1973) Pac Insects Monogr 31: the fruit flies (Tephritidae-Diptera) of Thailand and bordering countries. Entomology Department, Bernice P. Bishop Museum, Honolulu, pp 1–353
Harpending HC, Batzer MA, Gurven M, Jorde LB, Rogers AR et al (1998) Genetic traces of ancient demography. Proc Natl Acad Sci USA 95:1691–1697
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BMC Genet 6:13. v.3.23. http://ibdws.sdsu.edu/
Kapoor VC (1993) Indian fruit flies (Insecta: Diptera: Tephritidae). Oxford & IBH Publishing Co Pvt Ltd, New Delhi. ISBN 81-204-0745-8
Krafsur ES (2005) Role of population genetics in the sterile insect technique. In: Dyck VA, Hendrichs J, Robinson AS (eds) Sterile insect technique: principles and practice in area-wide integrated pest management. Springer, Dordrecht, pp 389–406
Li W, Yang L, Tang K, Zeng L, Liang G (2007) Microsatellite polymorphism of Bactrocera dorsalis (Hendel) populations in China. Acta Entomol Sin 50(12):1255–1262
Li Y, Li Z, Wu G, Wang H, Deng Y, Gong X (2009) A primary on the population genetics relationship of Bactrocera dorsalis (Diptera: Tephritidae) based on mtDNA ND6 gene. Biol Control Chin J 42–50. http://www.biomedcentral.com/sfx_links?ui=1471-2148-14-55&bibl=B12
Li Y, Wu Y, Chen H, Wu J, Li Z (2012) Population structure and colonization of Bactrocera dorsalis (Diptera: Tephritidae) in China, inferred from mtDNA COI sequences. J Appl Entomol 136(4):241–251
Liang F, Wu JJ, Liang GQ (2001) The first report of the test on the flight ability of oriental fruit fly. Acta Agric Univ Jiangxi 2:259–260
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452
Lombaert E, Guillemaud T, Cornuet JM, Malausa T, Facon B, Estoup A (2010) Bridgehead effect in the worldwide invasion of the biocontrol harlequin ladybird. PLoS One 5:e9743
Lunt DH, Zhang DX, Szymura JM, Hewitt GM (1996) The insect cytochrome oxidase I gene: evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol Biol 5:153–165
Lux SA, Copeland RS, White IM et al (2003) A new invasive fruit fly species from the Bactrocera dorsalis (Hendel) group detected in East Africa. Insect Sci Appl 23:355–361
Lyra ML, Fresia P, Gama S, Cristina J, Klaczko LB, Azeredo-Espin AML (2005) Analysis of mitochondrial DNA variability and genetic structure in populations of New World screwworm flies (Diptera: Calliphoridae) from Uruguay. J Med Entomol 42:589–595
Mantel N (1967) The detection of disease and a generalized regression approach. Cancer Res 27:209–220
Miller NJ, Dorhout DL, Rice ME, Sappington TW (2009) Mitochondrial DNA variation and range expansion in western bean cutworm (Lepidoptera: Noctuidae): no evidence for a recent population bottleneck. Environ Entomol 1:274–280
Mun J, Bohonak AJ, Roderick GK (2003) Population structure of the pumpkin fruit fly Bactrocera depressa (Tephritidae) in Korea and Japan: Pliocene allopatry or recent invasion? Mol Ecol 12:2941–2951
Nardi F, Carapelli A, Dallai R, Roderick GK, Frati F (2005) Population structure and colonization history of the olive fly, Bactrocera oleae (Diptera, Tephritidae). Mol Ecol 14:2729–2738
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76(10):5269–5273
Prabhakar CS, Sood P, Kapoor V, Kanwar SS, Mehta PK, Sharma PN (2009) Molecular and biochemical characterization of three bacterial symbionts of fruit fly, Bactrocera tau (Tephritidae: Diptera). J Gen Appl Microbiol 55:213–220
Prabhakar CS, Mehta PK, Sood P, Singh SK, Sharma P, Sharma PN (2012a) Population genetic structure of the melon fly, Bactrocera cucurbitae (Coquillett) (Diptera: Tephritidae) based on mitochondrial cytochrome oxidase (COI) gene sequences. Genetica 140:83–91
Prabhakar CS, Sood P, Mehta PK (2012b) Fruit fly (Diptera: Tephritidae) diversity in cucurbit fields and surrounding forest areas of Himachal Pradesh, a North-Western Himalayan state of India. Arch Phytopath Plant Protect 45(10):1210–1217
Prabhakar CS, Sood P, Mehta PK, Sharma PN (2013) Population genetic structure of the pumpkin fruit fly, Bactrocera tau (Walker) (Diptera: Tephritidae) in Himachal Pradesh, India. Biochem Syst Ecol 51:291–296
Roderick GK (1996) Geographic structure of insect populations: gene flow, phylogeography, and their uses. Annu Rev Entomol 41:263–290
Roderick GK, Navajas M (2003) Genes in new environments: genetics and evolution in biological control. Nat Rev Genet 4:889–899
Rogers A, Harpending H (1992) Population growth makes waves in the distribution of pairwise differences. Mol Biol Evol 9:552–569
Rollins LA, Woolnough AP, Sinclair R, Mooney NJ, Sherwin WB (2011) Mitochondrial DNA offers unique insights into invasion history of the common starling. Mol Ecol 20:2307–2317
Rosetti N, Remis MI (2012) Spatial genetic structure and mitochondrial DNA phylogeography of argentinean populations of the grasshopper Dichroplus elongatus. PLoS One 7(7):e40807. doi:10.1371/journal.pone.0040807
Schutze MK, Mahmood K, Pavasovic A, Wang BO, Newman J, Clarke AR, Krosch MN, Cameron SL (2014) One and the same: integrative taxonomic evidence that the African Invasive Fruit Fly Bactrocera invadens (Diptera: Tephritidae), is the same species as the oriental fruit fly Bactrocera dorsalis. Syst Entomol. doi:10.1111/syen.12114
Schutze MK, Aketarawong N, Amornsak W, Armstrong KF, Augustinos AA, Barr NB, Bo W, Bourtzis K, Boykin LM et al (2015) Synonymization of key pest species within the Bactrocera dorsalis species complex (Diptera: Tephritidae): taxonomic changes based on a review of 20 years of integrative morphological, molecular, cytogenetic, behavioural and chemoecological data. Syst Entomol 40(2):456–471
Shi W, Kerdelhue C, Ye H (2005) Population genetics of the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae), in Yunnan (China) based on mitochondrial DNA sequences. Environ Entomol 34:977–983
Shi W, Kerdelhue C, Ye H (2010) Population genetic structure of the oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae) from Yunnan province (China) and nearby sites across the border. Genetica 138:377–385
Shi W, Kerdelhue C, Ye H (2012) Genetic structure and inferences on potential source areas for Bactrocera dorsalis (Hendel) based on mitochondrial and microsatellite markers. PLoS One 7(5):e37083. doi:10.1371/journal.pone.0037083
Siedchlag AC, Benozzati ML, Passoni JC, Rodrigues MT (2010) Genetic structure, phylogeny, and biogeography of Brazilian eyelid-less lizards of genera Calyptommatus and Nothobachia (Squamata, Gymnophthalmidae) as inferred from mitochondrial DNA sequences. Mol Phylogenet Evol 2:622–630
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87(6):651–701
Slatkin M (1985) Gene flow in natural populations. Ann Rev Ecol Syst 16:393–430
Slatkin M, Hudson RR (1991) Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129:555–562
Sridhar V, Verghese A, Vinesh LS, Jayashankar M, Kamala Jayanthi PD (2014) CLIMEX simulated predictions of oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae) geographical distribution under climate change situations in India. Curr Sci 106(12):1702–1710
Stephens AEA, Kriticos DJ, Leriche A (2007) The current and future potential geographical distribution of the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Bull Entomol Res 97:369–378
Stonehouse JM (2001) An overview of fruit fly research knowledge and needs in the Indian Ocean region. In: Proceedings of the second national symposium on integrated pest management (IPM) in horticultural crops: new molecules, biopesticides and environment, Bangalore
Suare NM, Betancor E, Klassert TE, Almeida T, Hernandez M (2009) Phylogeography and genetic structure of the Canarian common chaffinch (Fringilla coelebs) inferred with mtDNA and microsatellite loci. Mol Phylogenet Evol 2:556–564
Suarez AV, Tsutsui ND (2008) The evolutionary consequences of biological invasions. Mol Ecol 17:351–360
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 3:585–595
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Torres TT, Lyra ML, Fresia P, Azeredo-Espin AML (2007) Assessing genetic variation in New World Screwworm Cochliomyia hominivorax populations from Uruguay. In: Vreysen MJB, Robinson AS, Hendrichs J (eds) Area-wide control of insect pests: from research to field implementation. Springer, Dordrecht, pp 183–191
Venkatesan M, Westbrook CJ, Hauer MC, Rasgon JL (2007) Evidence for a population expansion in the West Nile virus vector Culex tarsalis. Mol Biol Evol 5:1208–1218
Verghese A, Jayanthi PDK (2001) Integrated pest management in fruits. In: Parvatha Reddy P, Verghese A, Krishna Kumar NK (eds) Pest management in horticultural ecosystems. Capital Publishing Company, New Delhi, pp 1–23
Wan X, Nardi F, Zhang B, Liu Y (2011) The oriental fruit fly, Bactrocera dorsalis, in China: origin and gradual inland range expansion associated with population growth. PLoS One 6(10):e25238. doi:10.1371/journal.pone.0025238
Wan X, Liu Y, Zhang B (2012) Invasion history of the oriental fruit fly, Bactrocera dorsalis, in the Pacific-Asia region: two main invasion routes. PLoS One 7(5):e36176. doi:10.1371/journal.pone.0036176
Wilson JRU, Dormontt EE, Prentis PJ, Lowe AJ, Richardson DM (2009) Something in the way you move: dispersal pathways affect invasion success. Trends Ecol Evol 24:136–144
Wu ZZ, Li HM, Bin SY, Ma J et al (2014) Sequence analysis of mitochondrial ND1 gene can reveal the genetic structure and origin of Bactrocera dorsalis s.s. BMC Evol Biol 14:55
Yu DJ, Xu L, Nardi F, Li JG, Zhang RJ (2007) The complete nucleotide sequence of the mitochondrial genome of the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Gene 396:66–74. doi:10.1016/j.gene.2007.02.023
Acknowledgments
We thank Dr. Mukesh Kumar (Gujarat), Sh. Shravan Naga (Rajasthan), Dr. S. K. Naik (Bihar) and Sh. Pramod Kumar (Odisha & Uttar Pradesh) for assistance in collection of samples from respective sites spanning over India. We also thank to Dr. Pankaj Sood, Department of Entomology, CSK Himachal Pradesh Agricultural University, Palampur, Himachal Pradesh, India, Dr. Santosh S. Mali, ICAR-RCER, Research Centre, Plandu, Ranchi, Jharkhand, India and anonymous reviewers for their constructive comments and editing of final manuscript. We are grateful to Dr. B.P. Bhatt (Director of institute), Dr. A.K. Singh (Head of centre) and Dr. Bikash Das (Project leader) for giving valuable suggestions and providing laboratory facilities. This research was funded by Ministry of Agriculture, Government of India through National Initiative on Climate Resilient Agriculture (NICRA) project under Indian Council of Agricultural Research (ICAR) (ICAR-RCER/RC R/E.F./2011/29). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Choudhary, J.S., Naaz, N., Prabhakar, C.S. et al. Genetic analysis of oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae) populations based on mitochondrial cox1 and nad1 gene sequences from India and other Asian countries. Genetica 144, 611–623 (2016). https://doi.org/10.1007/s10709-016-9929-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-016-9929-7