Abstract
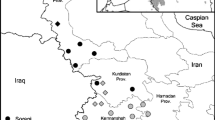
Past and recent climatic changes induced shifts in species ranges. Mantis religiosa has also expanded its range across Germany within the past decades. To determine the ancestry of German M. religiosa we sequenced four mitochondrial genes (COI, COII, Cyt b, ND4) of European M. religiosa populations. We found an east, central and west European lineage of M. religiosa. These distinct lineages are consistent with genetic isolation by distance during glacial periods, and the re-colonization of northern parts of Europe by species from different refugia. Within Germany, we found haplotypes clustering to the central and west European lineage suggesting that M. religiosa immigrated from two directions into Germany. Mismatch distributions, and negative Tajima’s D and Fu’s Fs values indicate a current range expansion of the central and west European lineage. We hypothesise that ongoing global warming which increases the availability of thermally favourable areas in Germany for M. religiosa adds to its current range expansion. In conclusion, M. religiosa colonized Germany via two directions: west German populations descended from French populations and east German populations from Czech populations.




Similar content being viewed by others
References
Akaike H (1979) A bayesian extension of the minimum AIC procedure of autoregressive model fitting. Biometrika 66:237–242
Aspöck H (2008) Postglacial formation and fluctuations of the biodiversity of Central Europe in the light of climate change. Parasitol Res 103:7–10
Badeck F, Pompe S, Kühn I, Glauer A (2008) Wetterextreme und Artenvielfalt. Zeitlich hochauflösende Klimainformationen auf dem Messtischblattraster und für Schutzgebiete in Deutschland. Naturschutz und Landschaftsplanung 40:343–345
Ballard JWO, Kreitman M (1995) Is mitochondrial DNA a strictly neutral marker? Trends Ecol Evol 10:485–488
Barr NB (2009) Pathway analysis of Ceratitis captitata (Diptera: Tephritidae) using mitochondrial DNA. J Econ Entomol 102:401–411
Berg MK, Schwarz CJ, Mehl JE (2011) Die Gottesanbeterin. Westarp Wissenschaften, Hohenwarsleben
Brechtel F (1996) Neozoen: neue Insektenarten in unserer Natur? In: Kinzelbach R, Schmidt-Fischer S (eds) Gebhardt H. Gebietsfremde Tierarten. ecomed, Landsberg, pp 127–154
Bull JJ, Huelsenbeck JP, Cunningham CW, Swofford DL, Waddell PJ (1993) Partitioning and combining data in phylogenetic analysis. Syst Biol 42:384–397
Bylebyl K, Poschlod P, Reisch C (2008) Genetic variation of Eryngium campestre L. (Apiaceae) in Central Europe. Mol Ecol 17:3379–3388
Cooper SJB, Ibrahim KM, Hewitt GM (1995) Postglacial expansion and genome subdivision in the European grasshopper Chorthippus parallelus. Mol Ecol 4:49–60
Detzel P (1995) Herkunft und Verbreitung der Heuschrecken in Baden-Württemberg. Articulata 10:107–118
Detzel P, Ehrmann R (1998) Mantis religiosa. In: Detzel P (ed) Die Heuschrecken Baden-Württembergs. Ulmer, Stuttgart (Hohenheim), pp 181–187
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Ehrmann R (1985) Standorttreue von Mantis religiosa (L.). Articulata 2:179–180
Ehrmann R (2003) Die Gottesanbeterin (Mantis religiosa), Neufunde in Deutschland. Articulata 18:253–254
Excoffier L (2004) Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol Ecol 13:853–864
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Fu Y (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Fu Y, Li W (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Hammouti N, Schmitt T, Seitz A, Kosuch J, Veith M (2010) Combining mitochondrial and nuclear evidences: a refined evolutionary history of Erebia medusa (Lepidoptera: Nymphalidae: Satyrinae) in Central Europe based on the COI gene. J Zool Syst Evol Res 48:115–125
Harpending HC (1994) Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol 66:591–600
Harz K (1957) Die Gottesanbeterin. Natur und Volk 87:187–193
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Hewitt GM (1999) Post-glacial re-colonization of European biota. Biol J Linn Soc 68:87–112
Hickling R, Roy DB, Hill JK, Fox R, Thomas CD (2006) The distribution of a wide range of taxonomic groups are expanding polewards. Glob Change Biol 12:450–455
Hideg JI (1994) The territorial behaviour of the Mantis religiosa and its migration propensity. Bull inf Soc Lepid Rom 5:291–296
Hideg JI (1996) Imbalances between the sexes in Mantis religiosa populations. Entomol Romanica 1:77–82
Li T, Zhang M, Qu Y, Ren Z, Zhang J, Guo Y et al (2011) Population genetic structure and phylogeographical pattern of rice grasshopper, Oxya hyla intricata, across Southeast Asia. Genetica 139:511–524
Liana A (2007) Distribution of Mantis religiosa (L.) and its changes in Poland. Fragmenta Faunistica 50:91–125
Lunt DH, Ibrahim KM, Hewitt GM (1998) mtDNA phylogeography and postglacial patterns of subdivision in the meadow grasshopper Chorthippus parallelus. Heredity 80:633–641
Parmesan C (2006) Ecological and evolutionary responses to recent climate change. Annu Rev Ecol Evol Syst 37:637–669
Pulliam RH (2000) On the relationship between niche and distribution. Ecol Lett 3:349–361
Reis DM, Cunha RL, Patrao C, Rebelo R, Castilho R (2011) Salamandra salamandra (Amphibia: Caudata: Salamandridae) in Portugal: not all black and yellow. Genetica 139:1095–1105
Robinet C, Roques A (2010) Direct impacts of recent climate warming on insect populations. Integr Zool 5:132–142
Rogers AR, Harpending HC (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Root TL, Price JT, Hall KR, Schneider SH, Rosenzweig C, Pounds AJ (2003) Fingerprints of global warming on wild animals and plants. Nature 421:57–60
Salt RW, James HG (1947) Low temperature as a factor in the mortality of eggs of Mantis religiosa L. Can Entomol 79:33–36
Scataglini MA, Lanteri AA, Confalonieri VA (2006) Diversity of boll weevil populations in South America: a phylogeographic approach. Genetica 126:353–368
Swofford DL (2003) PAUP*. Sinauer Associates, Sunderland
Taberlet P, Fumagalli L, Wust-Saucy A, Cosson J (1998) Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7:453–464
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tajima F (1996) The amount of DNA polymorphism maintained in a finite population when the neutral mutation rate varies among sites. Genetics 143:1457–1465
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol 9:678–687
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Walther G, Post E, Convey P, Menzel A, Parmesan C, Beebee TJC et al (2002) Ecological responses to recent climate change. Nature 416:389–395
Zitari-Chatti R, Chatti N, Fulgione D, Caiazza I, Aprea G, Elouaer A et al (2009) Mitochondrial DNA variation in the caramote prawn Penaeus (Melicertus) kerathurus across a transition zone in the Mediterranean Sea. Genetica 136:439–447
Acknowledgments
We are extremely grateful to Roberto Battiston, Manfred Berg, Enrico Busato, Claudine Decourchelle, Denis Loupy, Reinhard Ehrmann, Sönke Hardersen, Manfred Keller, Ingmar Landeck, Nora Lieskonig and Harald Krenn, Thomas Michaelis, Luca Picciau, Gerhard Pohl, Susanne Randolf, Ralf Rasch, Kai Schütte, and Christopher Tuchscherer for collecting and/or providing samples of M. religiosa. We also thank Christiane Groß for DNA preparation and Jes Johannesen for precious advice in data evaluation. We are especially grateful to Petr Janšta for providing genetic data from his phylogeographic database on M. religiosa. Additionally we thank Rebecca Nagel for linguistic improvement and the members of our working group, as well as the two anonymous reviewers for their valuable comments on an earlier version of this manuscript. This research was supported by a grant from the Deutsche Bundesstiftung Umwelt. This paper is part of the PhD thesis of Catherine Anne Linn.
Conflict of interest
The authors declare no conflict of interest.
Ethical standard
This article does not contain any experiments with animals performed by any of the authors. For collecting M. religiosa tissue samples we got the permissions from the nature conservation authorities.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Linn, C.A., Griebeler, E.M. Reconstruction of two colonisation pathways of Mantis religiosa (Mantodea) in Germany using four mitochondrial markers. Genetica 143, 11–20 (2015). https://doi.org/10.1007/s10709-014-9806-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-014-9806-1