Abstract
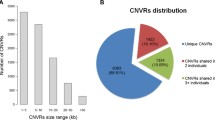
Copy number variation (CNV) has been recently examined in many species and is recognized as being a source of genetic variability, especially for disease-related phenotypes. In this study, the PennCNV software, a genome-wide CNV detection system based on the 60 K SNP BeadChip was used on a total sample size of 1,310 Beijing-You chickens (a Chinese local breed). After quality control, 137 high confidence CNVRs covering 27.31 Mb of the chicken genome and corresponding to 2.61 % of the whole chicken genome. Within these regions, 131 known genes or coding sequences were involved. Q-PCR was applied to verify some of the genes related to disease development. Results showed that copy number of genes such as, phosphatidylinositol-5-phosphate 4-kinase II alpha, PHD finger protein 14, RHACD8 (a CD8α- like messenger RNA), MHC B-G, zinc finger protein, sarcosine dehydrogenase and ficolin 2 varied between individual chickens, which also supports the reliability of chip-detection of the CNVs. As one source of genomic variation, CNVs may provide new insight into the relationship between the genome and phenotypic characteristics.


Similar content being viewed by others
References
Abernathy J, Li X, Jia X, Chou W, Lamont SJ, Crooijmans R (2014) Copy number variation in Fayoumi and Leghorn chickens analyzed using array comparative genomic hybridization. Anim Genet 45(3):400–411
Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, Hodges MD, Bhangal G, Patel SG, Sheehan-Rooney K, Duda M, Cook PR, Evans DJ, Domin J, Flint J, Boyle JJ, Pusey CD, Cook HT (2006) Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439:851–855
Bacon LD, Witter RL, Crittenden LB, Fadly A, Motta J (1981) B-Haplotype influence on Marek’s disease, Rous sarcoma, and lymphoid leucosis virus-induced tumors in chickens. Poult Sci 60:1132–1139
Bacon LD, Hunt HD, Cheng HH (2001) Genetic resistance to Marek’s disease. Curr Top Microbiol Immunol 255:121–141
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Beckmann JS, Estivill X, Antonarakis SE (2007) Copy number variants and gengetic traits: closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet 8:639–646
Bickhart DM, Hou Y, Schroeder SG, Alkan C, Cardone MF, Matukumalli LK, Song J, Schnabel RD, Ventura M, Taylor JF, Garcia JF, Van Tassel CP, Sonstegard TS, Eichler EE, Liu GE (2012) Copy number variation of individual cattle genomes using next-generation sequencing. Genome Res 22(4):778–790
Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M (2003) A proteomics strategy to elucidate functional protein–protein interactions applied to EGF signaling. Nat Biotechnol 21:315–318
Caricasole A, Duarte A, Larsson SH, Hastie ND, Little M, Holmes G, Todorov I, Ward A (1996) RNA binding by the Wilms tumor suppressor zinc finger proteins. Proc Natl Acad Sci USA 93:7562–7566
Crooijmans R, Fife M, Fitzgerald T, Strickland S, Cheng H, Kaiser P, Redon R, Groenen M (2013) Large scale variation in DNA copy number in chicken breeds. BMC Genomics 14:398
Dellinger AE, Saw SM, Goh LK, Seielstad M, Young TL, Li YJ (2010) Comparative analyses of seven algorithms for copy number variant identification from single nucleotide polymorphism arrays. Nucleic Acids Res 38(9):e105
Doody GM, Billadeau DD, Clayton E, Hutchings A, Berland R, McAdam S, Leibson PJ, Turner M (2000) Vav-2 controls NFAT-dependent transcription in B- but not T-lymphocytes. EMBO J 19(22):6173–6184
Dorshorst BJ, Siegel PB, Ashwell CM (2010) Genomic regions associated with antibody response to sheep red blood cells in the chicken. Anim Genet 42:300–308
Elferink MG, Vallée AA, Jungerius AP, Crooijmans RP, Groenen MA (2008) Partial duplication of the PRLR and SPEF2 gene at the late feathering locus in chicken. BMC Genomics 9:391
Fadista J, Nygaard M, Holm LE, Thomsen B, Bendixen C (2008) A snapshot of CNVs in the pig genome. PLoS One 3(12):e3916
Feuk L, Carson AR, Scherer SW (2006) Structural variation in the human genome. Nat Rev Genet 7:85–97
Fiegler H, Redon R, Andrews D, Scott C, Andrews R, Carder C, Clark R, Dovey O, Ellis P, Feuk L, French L, Hunt P, Kalaitzopoulos D, Larkin J, Montgomery L, Perry GH, Plumb BW, Porter K, Rigby RE, Rigler D, Valsesia A, Langford C, Humphray SJ, Scherer SW, Lee C, Hurles ME, Carter NP (2006) Accurate and reliable high-throughput detection of copy number variation in the human genome. Genome Res 16:1566–1574
Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Aburatani H, Jones KW, Tyler-Smith C, Hurles ME, Carter NP, Scherer SW, Lee C (2006) Copy number variation: new insights in genome diversit. Genome Res 16:949–961
Glessner JT, Wang K, Cai GQ, Korvatska O, Kim CE, Wood S, Zhang HT, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PMA, Chiavacci R, Annaiah K, Thomas K, Hou CP, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T, Game RM, Rudd DS, Zurawiecki D, McDougle CJ, Davis LK, Miller J, Posey DJ, Michaels S, Kolevzon A, Silverman JM, Bernier R, Levy SE, Schultz RT, Dawson G, Owley T, McMahon WM, Wassink TH, Sweeney JA, Nurnberger JI, Coon H, Sutcliffe JS, Minshew NJ, Grant SFA, Bucan M, Cook EH, Buxbaum JD, Devlin B, Schellenberg GD, Hakonarson H (2009) Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459:569–573
Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, Murthy KK, Rovin BH, Bradley W, Clark RA, Anderson SA, O’Connell RJ, Agan BK, Ahuja SS, Bologna R, Sen L, Dolan MJ, Ahuja SK (2005) The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307:1434–1440
Gou ZY, Liu RR, Zhao GP, Zheng MQ, Li P, Wang HH, Zhu Y, Chen JL, Wen J (2012) Epigenetic modification of TLRs in leukocytes is associated with increased susceptibility to Salmonella enteritidis in chickens. PLoS One 7:e33627
Griffin DK, Robertson LB, Tempest HG, Vignal A, Fillon V, Crooijmans RP, Groenen MA, Deryusheva S, Gaginskaya E, Carré W, Waddington D, Talbot R, Völker M, Masabanda JS, Burt DW (2008) Whole genome comparative studies between chicken and turkey and their implications for avian genome evolution. BMC Genomics 9:168
Guryev V, Saar K, Adamovic T, Verheul M, Van Heesch C, Cook S, Pravenec M, Aitman T, Jacob H, Shull JD, Hubner N, Cuppen E (2008) Distribution and functional impact of DNA copy number variation in the rat. Nat Genet 40:538–545
Hoang VT, Nguyen LT, Le HS, Eman AO, Thomas CB, Peter GK, Jurgen FJK, Velavan TP (2011) Ficolin-2 levels and FCN2 haplotypes influence hepatitis B infection outcome in Vietnamese patients. PLoS One 6(11):e28113
Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U (2006) A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet 38:1173–1177
Ionita-Laza I, Rogers AJ, Lange C, Raby BA, Lee C (2009) Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis. Genomics 93:22–26
Liu X, Jiang L, Wang A, Yu J, Shi F, Zhou X (2009) MicroRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Lett 286:217–222
Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM (2009) Finding the missing heritability of complex diseases. Nature 461:747–753
McCarroll SA (2008) Extending genome-wide association studies to copy-number variation. Hum Mol Genet 17:R135–R142
Metzger J, Philipp U, Lopes SM, Machado CA, Felicetti M, Silvestrelli M, Dist O (2013) Analysis of copy number variants by three detection algorithms and their association with body size in horses. BMC Genomics 14:487
Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen WW, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang FT, Zhang JJ, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME (2006) Global variation in copy number in the human genome. Nature 444:444–454
Schwab SG, Knapp M, Sklar P, Eckstein GN, Sewekow C, Borrmann-Hassenbach M, Albus M, Becker T, Hallmayer JF, Lerer B, Maier W, Wildenauer DB (2006) Evidence for association of DNA sequence variants in the phosphatidylinositol-4-phosphate 5-kinase IIalpha gene (PIP4K2A) with schizophrenia. Mol Psychiatry 11(9):837–846
She XW, Cheng Z, Zollner S, Church DM, Eichler EE (2008) Mouse segmental duplication and copy number variation. Nat Genet 40:909–914
Skinner BM, Robertson LB, Tempest HG, Langley EJ, Ioannou D, Fowler KE, Crooijmans RP, Hall AD, Griffin DK, Volker M (2009) Comparative genomics in chicken and Pekin duck using FISH mapping and microarray analysis. BMC Genomics 10:357
Stankiewicz P, Lupski JR (2010) Structural variation in the human genome and its role in disease. Annu Rev Med 61(1):437–455
Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, Tyler-Smith C, Carter N, Scherer SW, Tavare S, Deloukas P, Hurles ME, Dermitzakis ET (2007) Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315:848–853
Takako A, Kohichiroh Y, Yasuyuki G, Nobuhisa Y, Akira T, Osamu D, Hironori M, Nobuaki Y, Yoshito I, Yuji N, Toshikazu Y (2013) Aberrant expression of the PHF14 gene in biliary tract cancer cells. Oncol Lett 5:1849–1853
Tian M, Wang YQ, Gu XR, Feng CG, Fang SY, Hu XX, Li N (2013) Copy number variants in locally raised Chinese chicken genomes determined using array comparative genomic hybridization. BMC Genomics 14:262
Volker M, Backstrom N, Skinner BM, Langley EJ, Bunzey SK, Ellegren H, Griffin DK (2010) Copy number variation, chromosome rearrangement, and their association with recombination during avian evolution. Genome Res 20:503–511
Wang XF, Byers S (2014) Copy number variation in Chickens: a review and future prospects. Microarrays 3:24–38
Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M (2007) PennCNV: an integrated hidden Markov model designed for highresolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 17(11):1665–1674
Wang XF, Nahashon S, Feaster TK, Bohannon-Stewart A, Adefope N (2010) An initial map of chromosomal segmental copy number variations in the chicken. BMC Genomics 3(11):351
Wang JY, Jiang JC, Fu WX, Li J, Ding XD, Liu JF, Zhang Q (2012a) A genome-wide detection of copy number variations using SNP genotyping arrays in swine. BMC Genomics 13:273
Wang Y, Gu X, Feng C, Song C, Hu X, Li N (2012b) A genome-wide survey of copy number variation regions in various chicken breeds by array comparative genomic hybridization method. Anim Genet 43(3):282–289
Wu CM, Zhao GP, Wen J, Chen JL, Zheng MQ, Chen GH (2007) Diversity of immune traits between Chinese Beijing-You chicken and White Leghorn. Acta Veterinaria et Zootechnica S inica 38(12):1383–1388 (In Chinese)
Zhang F, Gu WL, Hurles ME, Lupski JR (2009) Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 10:451–481
Acknowledgments
The authors thank Dr. W. Bruce Currie (Emeritus Professor, Cornell University, USA) for help with this manuscript. The work was supported by the National Key Technology R&D Program (2011BAD28B03), and the National Nonprofit Institute Research Grant (2012ZL068, 2011CJ-6).
Author information
Authors and Affiliations
Corresponding author
Additional information
Wei Zhou and Ranran Liu, have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10709_2014_9788_MOESM1_ESM.doc
The supplementary materials of this paper are shown in Table S (Primers, the Novel CNVRs from this study and those previously found, ENSEMBL_GENE_ID, genes compared in the DGV database)
Rights and permissions
About this article
Cite this article
Zhou, W., Liu, R., Zhang, J. et al. A genome-wide detection of copy number variation using SNP genotyping arrays in Beijing-You chickens. Genetica 142, 441–450 (2014). https://doi.org/10.1007/s10709-014-9788-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-014-9788-z