Abstract
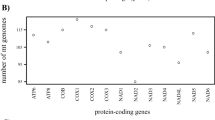
While it is well known that changes in the general processes of molecular evolution have occurred on a variety of timescales, the mechanisms underlying these changes are less well understood. Parasitic lice (“Phthiraptera”) and their close relatives (infraorder Nanopsocetae of the insect order Psocodea) are a group of insects well known for their unusual features of molecular evolution. We examined changes in base composition across parasitic lice and bark lice. We identified substantial differences in percent GC content between the clade comprising parasitic lice plus closely related bark lice (=Nanopsocetae) versus all other bark lice. These changes occurred for both nuclear and mitochondrial protein coding and ribosomal RNA genes, often in the same direction. To evaluate whether correlations in base composition change also occurred within lineages, we used phylogenetically controlled comparisons, and in this case few significant correlations were identified. Examining more constrained sites (first/second codon positions and rRNA) revealed that, in comparison to the other bark lice, the GC content of parasitic lice and close relatives tended towards 50 % either up from less than 50 % GC or down from greater than 50 % GC. In contrast, less constrained sites (third codon positions) in both nuclear and mitochondrial genes showed less of a consistent change of base composition in parasitic lice and very close relatives. We conclude that relaxed selection on this group of insects is a potential explanation of the change in base composition for both mitochondrial and nuclear genes, which could lead to nucleotide frequencies closer to random expectation (i.e., 50 % GC) in the absence of any mutation bias. Evidence suggests this relaxed selection arose once in the non-parasitic common ancestor of Phthiraptera + Nanopsocetae and is not directly related to the evolution of the parasitism in lice.


Similar content being viewed by others
References
Cameron D, Crespi BJ (1995) Do long branches attract flies? Nature 373:666
Cameron SL, Johnson KP, Whiting MF (2007) The mitochondrial genome of the screamer louse Bothriometopus (Phthiraptera: Ischnocera): effects of extensive gene rearrangements on the evolution of the genome as a whole. J Mol Evol 65:589–604
Cameron SL, Yoshizawa K, Mizukoshi A, Whiting MF, Johnson KP (2011) Mitochondrial genome deletions and minicircles are common in lice (Insecta: Phthiraptera). BMC Genom 12:394
Chiusano ML, D’Onofrio G, Alvarez-Valin F, Jabbari K, Colonna G, Bernardi G (1999) Correlations of nucleotide substitution rates and base composition of mammalian coding sequences with protein structure. Gene 238:23–31
Clark MA, Moran NA, Baumann P (1999) Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol Biol Evol 16:1586–1598
Dowton M, Austin AD (1995) Increased genetic diversity in mitochondrial genes is correlated with the evolution of parasitism in the Hymenoptera. J Mol Evol 16:298–309
Dowton M, Cameron SL, Dowavic JI, Austin AD, Whiting MF (2009) Characterisation of 66 mitochondrial gene rearrangements in the Hymenoptera reveal underlying trends in mitochondrial genome evolution. Mol Biol Evol 26:1607–1617
Felsenstein J (1978) Case in which parsimony or compatibility methods will be positively misleading. Syst Zool 27:401–410
Felsenstein J (1985) Phylogenies and the comparative method. Am Nat 125:1–15
Foster PG, Jermiin LS, Hickey DA (1997) Nucleotide composition bias affects amino acid content in protein coded by animal mitochondria. J Mol Evol 44:282–288
Fukatsu T, Koga R, Smith WA, Tanaka K, Nikoh N, Sasaki-Fukatsu K, Yoshizawa K, Dale C, Clayton DH (2007) Bacterial endosymbiont of the slender pigeon louse, Columbicola columbae, allied to endosymbionts of grain weevils and tsetse flies. App Env Microbiol 72:6660–6668
Galtier N, Gouy M (1995) Inferring phylogenies from DNA sequences of unequal base compositions. Proc Natl Acad Sci USA 92:11317–11321
Hafner MS, Sudman PD, Villablanca FX, Spradling TA, Demastes JW, Nadler SA (1994) Disparate rates of molecular evolution in cospeciating hosts and parasites. Science 265:1087–1090
Huelsenbeck JP (1997) Is the Felsenstein zone a fly trap? Syst Biol 46:69–74
Jermiin LS, Graur D, Lowe RM, Crozier RH (1994) Analysis of directional mutation pressure and nucleotide content in mitochondrial cytochrome b genes. J Mol Evol 39:160–173
Johnson KP, Seger J (2001) Elevated rates of nonsynonymous substitution in island birds. Mol Biol Evol 18:874–881
Johnson KP, Cruikshank RH, Adams RJ, Smith VS, Page RDM, Clayton DH (2003) Dramatically elevated rates of mitochondrial substitution in lice (Insecta: Phthiraptera). Mol Phylog Evol 26:231–242
Johnson KP, Yoshizawa K, Smith VS (2004) Multiple origins of parasitism in lice. Proc R Soc Lond B 271:1771–1776
Johnson KP, Walden KKO, Robertson HM (2013) Next-generation phylogenomics using a target restricted assembly method. Mol Phylogenet Evol 66:417–422
Kambhampati S, Rai KS, Verleye DM (1992) Frequencies of mitochondrial DNA haplotypes in laboratory cage populations of the mosquito, Aedes albopictus. Genetics 132:205–209
Kimura M (1962) On the probability of fixation of mutant genes in a population. Genetics 47:713–719
Kirkness EF, Haas BJ, Sun W et al (2010) Genome sequence of the human body louse and primary endosymbiont provide insights into the permanent parasitic lifestyle. PNAS 107:12168–12173
Kosakovsky Pond SL, Frost SDW, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679
Leigh EG Jr (1970) Natural selection and mutability. Am Nat 104:301–305
Li W-H (1997) Molecular evolution. Sinauer Associates, Sunderland
Maddison DR, Maddison WP (2005) MacClade ver 4.08. Sinauer Associates, Sunderland
Maddison WP, Maddison DR (2010) Mesquite: a molecular system for evolutionary analysis. Ver. 2.74. http://mesquiteproject.org. Accessed 30 June 2011
Matsuo Y (2003) Evolution of the GC content of the histone 3 gene in seven Drosophila species. Genes Genet Syst 78:309–318
Midford PE, Garland Jr. T, Maddison WP (2010) PDAP package of Mesquite. Ver. 1.15. http://mesquiteproject.org/pdap_mesquite/index.html. Accessed 30 June 2011
Moriyama EN, Gojobori T (1992) Rates of synonymous substitution and base composition of nuclear genes in Drosophila. Genetics 130:855–864
Nigro L, Prout T (1990) Is there selection on RFLP differences in mitochondrial DNA? Genetics 125:551–555
Ohta T (1973) Slightly deleterious mutant substitutions in evolution. Nature 246:96–98
Ohta T (1992) The nearly neutral theory of molecular evolution. Ann Rev Ecol Syst 23:263–286
Page RDM, Lee PLM, Bocher SA, Griffiths R, Clayton DH (1998) A different tempo of mitochondrial DNA evolution in birds and their ectoparasitic lice. Mol Phylog Evol 9:276–293
Page RDM, Cruickshank R, Johnson KP (2002) Louse (Insecta: Phthiraptera) mitochondrial 12S rRNA secondary structure is highly variable. Insect Mol Biol 11:361–369
Rosenberg MS (ed) (2009) Sequence alignment. University of California Press, Barkley
SAS Institute Inc (2009) JMP, version 8. SAS Institution, Cray
Sasaki-Fukatsu K, Koga R, Nikoh N, Yoshizawa K, Kasai S, Mihara M, Kobayashi M, Tomita T, Fukatsu T (2006) Symbiotic bacteria associated with stomach discs of human lice. App Env Microbiol 72:7349–7352
Shao R, Campbell NJH, Barker SC (2001) Numerous gene rearrangements in the mitochondrial genome of wallaby louse, Heterodoxus macropus (Phthiraptera). Mol Biol Evol 18:858–865
Shao R, Kirkness EF, Barker SC (2009) The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse Pediculus humanus. Genome Res 19:904–912
Sharp PM, Li W-H (1989) On the rate of DNA sequence evolution in Drosophila. J Mol Biol 28:398–402
Sheffield NC, Song H, Cameron SL, Whiting MF (2009) Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst Biol 58:381–394
Shields DC, Sharp PM, Higgins DG, Wright F (1988) “Silent” sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol Biol Evol 5:704–716
Wei DD, Shao R, Yuan ML, Dou W, Barker SC, Wang JJ (2012) The multiparitite mitochondrial genome of Liposcelis bostrichophila: insights into the evolution of mitochondrial genomes in bilateral animals. PLoS ONE 7:e33973
Woolfit M, Bronham L (2003) Increased rates of sequence evolution in endosymbiotic bacteria and fungi with small effective population sizes. Mol Biol Evol 20:1545–1555
Yoshizawa K, Johnson KP (2003) Phylogenetic position of Phthiraptera (Insecta: Paraneoptera) and elevated rate of evolution in mitochondrial 12S and 16S rDNA. Mol Phylog Evol 29:102–114
Yoshizawa K, Johnson KP (2010) How stable is the “Polyphyly of Lice” hypothesis (Insecta: Psocodea)?: A comparison of phylogenetic signal in multiple genes. Mol Phylog Evol 55:939–951
Young ND, dePamphilis CW (2005) Rate variation in parasitic plants: correlated and uncorrelated patterns among plastid genes of different function. BMC Evol Biol 5:16
Acknowledgments
We thank C. Lienhard, E. L. Mockford, Y. Kamimura, L. Durden, and J. Klicka for specimens. We also thank Joe Felsenstein and Colin Dale for suggestions in data analyses. Thanks are also due to two anonymous reviewers for constructive comments that are helpful in improving the earlier ms. KY thanks E. Hasegawa and M. Ôhara for allowing the use of their molecular facilities. This study was supported by Japan Society for the Promotion of Science Grants 18770058 and 21770083 to KY and US National Science Foundation Grant DEB-0612938 to KPJ.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yoshizawa, K., Johnson, K.P. Changes in base composition bias of nuclear and mitochondrial genes in lice (Insecta: Psocodea). Genetica 141, 491–499 (2013). https://doi.org/10.1007/s10709-013-9748-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-013-9748-z