Abstract
Mobile elements using a “cut and paste” mechanism of transposition (Class II) are frequently prone to internal deletions and the question of the origin of these copies remains elusive. In this study, we looked for copies belonging to the Lemi Family (Tc1-mariner-IS630 SuperFamily) in the plant genomes, and copies within internal deletions were analyzed in detail. Lemi elements are found exclusively in Eudicots, and more than half of the copies have been deleted. All deletions occur between microhomologies (direct repeats from 2 to 13 bp). Copies less than 500 bp long, similar to MITEs, are frequent. These copies seem to result from large deletions occurring between microhomologies present within a region of 300 bp at both extremities of the element. These regions are particularly A/T rich, compared to the internal part of the element, which increases the probability of observing short direct repeats. Most of the molecular mechanisms responsible for double strand break repair are able to induce deletions between microhomologies during the repair process. This could be a quick way to reduce the population of active copies within a genome and, more generally, to reduce the overall activity of the element after it has entered a naive genome.
Similar content being viewed by others
Introduction
Although transposable elements (TEs) are present in almost all living organisms, few of them are active and the vast majority of copies are inactive. Their inactivation occurs as a result of mutations, truncations, insertions, or deletions. Mutations generally decrease the activity of the copy, and very few increase it. An example of the latter is the activity of the mos1 element (mariner Family), initially described in Drosophila mauritiana (Jacobson and Hartl 1985; Jacobson et al. 1986), which does not seems to be optimal, and artificial copies with greater activity can be constructed in the laboratory (Auge-Gouillou et al. 2001). Truncations (deletions including one or both ends of the element) result in copies that are unable to move, although some of them can produce functional transposition machinery usable by other copies. Insertions are frequent, particularly in regions where copies are inserted into each other, as reported in Zea mays (SanMiguel et al. 1996), in Fusarium oxysporum (Hua-Van et al. 2000) and more recently in a larger set of species (Gao et al. 2012). Finally, deletions (copies with internal deletion(s) and intact inverted terminal repeats—ITRs) are frequently observed, particularly in Class II elements. These copies cannot move alone, but are able to use the transposition machinery provided by other copies present in the same genome (trans-activation). Such copies are observed for all type of elements, but here we will focus solely on Class II elements.
Internal deletions can vary considerably in size, ranging from a few base pairs (bp) to several hundred bp, resulting in very variable copy lengths including MIniature Transposable Elements (MITEs), which have been detected in many genomes including plants, fungi, and insects (Casacuberta et al. 1998; Dufresne et al. 2007; Feschotte et al. 2002; Jiang et al. 2004) or in copies with short deletions like those belonging to the mauritiana SubFamily of mariner described in the Drosophilidae (Brunet et al. 2002).
Since non-autonomous copies (for many reasons mutations, insertions, deletions) are more frequent in genomes than full length and putatively active ones, the genesis and the dynamics are of interest. In this respect, the few reports of natural or experimental evolution dealing with the fate of full-length copies after their arrival or injection into a naive genome (i.e. a genome with no copies belonging to the same Family as that being injected), show that copies with internal deletions can emerge rapidly. For instance, KP copy seems to have emerged soon after the invasion of the P element in D. melanogaster (Anxolabehere et al. 1987; Daniels et al. 1987; Scavarda and Hartl 1987). Moreover, hobo with internal deletions were observed just a few generations after full-length copies had been injected into E strains (free of active hobo) of D. melanogaster (Galindo et al. 1995; Ladeveze et al. 1998). No such dynamics has been followed or developed in other species. Only theoretical approaches have been proposed showing that after the first step of genome invasion, the general activity of copies must decrease rapidly to avoid to lead to populations extinction (Le Rouzic et al. 2007). In plants, no experimental or natural observations (dynamics of an element just after its emergence or introduction into a naïve genome) have been done probably due to the longer generation time compared to species like Drosophila.
Two scenarios are generally proposed to explain the emergence of MITEs: internal deletion(s) and de novo emergence (Jiang et al. 2004). Arguments in favor of the first explanation are based on the similarities between deleted and full-length copies. However, because similarities are sometimes restricted to the ITR, the alternative explanation, involving de novo emergence, cannot be ruled out.
Internal deletions in Class II elements are mainly due to their transposition mechanism i.e. to “cut and paste”. The excision of these elements leaves a double strand break that has to be filled by a process of gap repair. In 1997, it was proposed that the gap repair mechanism, involved in the genesis of Ds non-autonomous copies from Ac autonomous ones in Z. mays, occurs through a synthesis-dependent, strand-annealing pathway, deletions being the result of “premature release of the newly synthesized single-strand DNA” leading to an Abortive Gap Repair (Rubin and Levy 1997). These authors also reported that recent deletions have been observed between two microhomologies. Brunet et al. (2002) also reported a similar observation from the analysis of more than 20 deleted copies of the mariner elements in Drosophilidae, where internal deletions occur between short direct repeats (SDR) of 5–8 bp. This was the first mechanism proposed to explain the emergence of internal deletions in Class II elements. However, today, other mechanisms can be evoked involving, for instance, a double strain break (DSB) inside a TE followed by precise or imprecise repair according to the mechanism concerned (NHEJ, MMEJ, SSA…). This will be discussed in the last part of this work.
In the de novo hypothesis, it is suggested that two almost identical sequences with opposite orientations, which are able to fix a transposase may occur by chance. If it does, these two inverted sequences and the sequence between them could be amplified by the transposase provided by a TE copy from elsewhere in the genome. Such types of sequences with opposite orientations have already been described in Saccharomyces cerevisiae (Achaz et al. 2000), suggesting that an active duplication mechanism resembling chromosome duplication and rearrangement could create close repeats. Because the binding site of the transposase is relatively short (about 10 bp), such a phenomenon demands serious consideration.
Interestingly, all elements do not have the same evolutionary trajectory since some of them seem to be more sensitive to internal deletions than others. Indeed, many MITEs are closely related to elements belonging to the Tc1-mariner-IS630 SuperFamily (Casacuberta et al. 1998; Dufresne et al. 2007; Jiang et al. 2004). Moreover, these deleted copies may be also more or less able to invade their genomic environment, and in some cases, only a few copies can be identified, while for others large numbers of copies (>100 or 1,000) are observed (see the previous references). However, in descriptive approaches (description of a genome at a t time), the ratio between full length and deleted copies probably depends on the timing of the deletion during the TE life cycle.
In the present work, a detailed analysis of the break points (BPs) of the deletions was carried out from a large set of TEs copies with internal deletions. These copies belong to the Lemi Family (Larger Emigrant) of the Tc1-mariner-IS630 SuperFamily. This Family was initially reported in Arabidopsis thaliana from MITEs copies (Casacuberta et al. 1998). Full-length copies providing the transposase were described 2 years later, suggesting that these MITEs derived from Pogo-like sequences (Feschotte and Mouches 2000). Later, new members of this Family were reported in Medicago truncatula (Guermonprez et al. 2008). Here, elements belonging to this Family were searched for using an in silico approach and were found in a large group of plants (all Eudicots). We show that most of the deletions occur between microhomologies and exactly at or near to the BPs.
Materials and methods
Detection of Lemi elements
To detect new copies of this Family, the sequences of AtLemi1 described from A. thaliana and of MtLemi1 from M. truncatula, were used as queries in Blast (with default parameters) in nucleotide databases (GeneBank http://srs.dna.affrc.go.jp/srs8/, but also from the sites of the Genome Project of several species: A. thaliana, A. lyrata, Glycine max and M. truncatula). When partial sequences were detected, i.e. segments shorter than the complete element of reference and without one or both extremities, the ITRs were searched for by an analysis of the flanking regions. In addition, searches were also carried out using consensus sequences of ITRs as queries. This led us to detect 4 categories of copies: 1—putative full-length copies (2,100–2,200 bp); 2—copies with insertions; 3—copies with internal deletions; 4—truncated copies lacking one or both extremities. Very few sequences (n = 5) belonging to this last category were found.
Classification
All copies were classified according to a previously described automatic method based on pairwise distances (Rouault et al. 2009). ClustalW (version 1.83) was used to align the sequences (manually checked) and distances were computed between all pairs of sequences, as the complement to 1 of identity (ratio of the number of nucleotides in identical positions over the total length of the alignment). Due to the existence of gaps in the alignments, the exact expression of the distance is: D = 1 − NI/(NS1 + wNS2), where NI is the number of positions where two identical nucleotides are observed, NS1 the total number of positions where two nucleotides are present, NS2 the number of positions for which a gap occurs facing a nucleotide and, w the gap weight (varying from 1 to 0 according to Rouault et al. 2009).
Analysis of deletions
Each sequence with an internal deletion was aligned against the reference sequence (GenBank acc no = AC141115 of M. truncatula, from nucleotide 28,902 to 31,902—reverse). Microhomologies were manually searched for by an exploration of the flanking regions (30 bp) of the deletion BPs, and only deletions of more than 5 bp were taken into consideration. A total of 181 deletions were analyzed in detail.
Results
Detection of Lemi elements
While Lemi elements were searched for without any filter, all of the 310 copies detected belong to the Eudicot clade (Fig. 1; Table 1). According to the classification of the Angiosperm Phylogeny Group, Eudicots are a non-monophyletic clade subdivided into four main groups (Group 2003; Soltis et al. 2000). The Lemi copies were found in species belonging to three of these groups, the rosids (Eurosids I and II) and asterids (Euasterids I).
At least two alternative scenarios could explain this uneven distribution (Capy et al. 1994): the existence of horizontal transfers between these species or the loss of Lemi elements from most of the species derived from the common ancestors of the core Eudicots. The fact that these elements are present in only four distantly-related orders out of the 29 orders of the core Eudicots argues in favor of the first scenario. However, we cannot exclude the alternative hypothesis (i.e. the loss of copies formerly present in a common ancestor). Indeed, the presence of copies belonging to several Tribes in A. thialiana (Table 1) could also be explained by the persistence of an ancestral polymorphism originating from a common ancestor. This is a perfect illustration of how cautious we must be in reaching our conclusions, all the more because complete genomes are not available for all species of Eudicots.
Classification and structure of Lemi elements
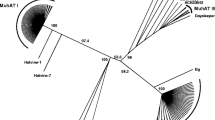
Lemi elements are members of the Tc1-mariner-IS630 SuperFamily, which is subdivided into several Families and SubFamilies mainly according to their sequence similarities (Wicker et al. 2007). Based on the automatic classification used, it is possible to identify about 18 Families within this SuperFamily, including the Lemi Family, which is closely related to the Pogo Family. The Lemi Family can then be subdivided into the Ogris and Poucetis SubFamilies (Fig. 2).
Members of the Ogris SubFamily have been detected in A. thaliana, Gossypium hirsutum and G. raimondii (Table 1), and include the full-length copy (AtLemi1 2,124 bp) reported in A. thaliana. The size of these 12 copies varies from 1,245 to 12,770 bp. The unusually large size of one copy observed in G. hirsutum (12,770 bp), is due to two insertions of partial copia elements (Grover et al. 2004). Initially, this copy was described as a member of the Pogo Family, but similarities clearly show that it is more closely related to the Lemi Family.
More sequences (n = 341) belonging to the Poucetis SubFamily have been discovered. Most of them are internally deleted, and very few seem to be complete, except in the Papilionis Tribe, which includes the complete copy reported in M. truncatula (Guermonprez et al. 2008). The size of these copies ranges from 301 to 5,777 bp, with a mean value of 1,048 bp. Figure 3 summarizes the distribution of the lengths of Ogris and Poucetis SubFamilies.
The papilionis Tribe
Copies belonging to the papilionis Tribe (including the MtLemi1 element of M. truncatula) are found in three species of the same order (Fabales): Lotus japonicus, Glycine max, and M. truncatula. It is quite possible that this distribution reflects the existence of these elements in the common ancestor. However, as previously pointed out, the absence of information about the genomes of all the species makes it impossible to conclude. However, the papilionis Tribe has the highest copy number, several of them being similar in size to the full-length reference copy (2,128 bp). A phylogeny performed on the longest copies (L > 2,035 bp) belonging to G. max and M. truncatula (the longest copy of L. japonicus was not included in this phylogeny: L = 1,416 bp) clearly shows that two specific groups can be identified, as they have a bootstrap value of 100 % between them (Figure 1 of supplementary data). It should also be pointed out that the branch lengths are shorter in the M. truncatula clade, suggesting that a more recent transposition burst has occurred in this species, as previously suspected by other authors (Guermonprez et al. 2008).
Classification of the Lemi Family within the SuperFamily Tc1-mariner-IS630. The main Families are mariner (Mar and Atl), Chlorophyllis (Chl or Plant mariner), Gambol (Gam), Tc1 (Tco), Matelotis (Mat or maT or Mori), Fot, Mosquitis (Msq or ITm-D37E) and Jap (IS630, ISRm10, IS870,…). According to the automatic classification (Rouault et al. 2009), the Lemi1 Family is split into two SubFamilies: Ogris (Ogr) and Poucetis (Pou). The SubFamily Ogris contains two Tribes: Barbetrouillis (Bar) and Shrekis (Shr). The SubFamily Poucetis contains a large majority of deleted sequences, and splits into ten Tribes: Papilionis (Pap), Solanis (Sol), Gobelinis (Gob), Hobgoblinis (Hob), Kobolnis (Kob), Gnomis (Gno), Lutinis (Lut), Ratlyris (Rat), Djinnis (Dji) and Korriganis (Kor). The arrows give the position of the two references sequences from A. thaliana (Barbetroullis Tribe—green) and from M. truncatula (Papilionis Tribe—brown)
Internal deletions of Lemi elements
Internal deletions were analyzed from 117 copies, 78 % of them from the Papilionis Tribes. The average number of deletions per copy is 1.5 and the median is 1 (74 copies with one deletion, 27 copies with two deletions, 12 with 3 deletions, 3 with 4 deletions and 1 with 5 deletions). The size distributions and locations of the deletions are given in Fig. 4. The deletion sizes of the copies belonging to the Papilionis Tribe ranged between 1 and 1,662 bp, with a tendency toward large deletions, while those of the members of the other Tribes range from 3 to 878 bp (Fig. 4a). Moreover, the part of the element susceptible to being deleted seems to be similar for the 5′ and 3′ ends (Fig. 4b), in contrast to what was observed for the mariner element of the mauritiana SubFamily (Brunet et al. 2002), in which a bias toward the 5′ end was observed.
The BPs of the deletions were then analyzed in detail from the sequences belonging to the Papilionis Tribe. It was always found that microhomologies were present exactly at the BP or in the flanking regions close to the BP. The events were divided into three categories: the presence of microhomologies exactly at the two BPs (BPEE 31 %), near the two BPs (BPNN 18 %), and one exactly at the BP and the other near the BP (BPEN 51 %). Some examples are given Fig. 5.
Examples of microhomologies observed at or close to the BPs of the deletions. BPEE: microhomologies are present exactly at the BP. BPNN: microhomologies are close to the BP. BPEN: one microhomology is present exactly at the BP, while the other is close to the BP. The right part of the Figure gives a schematic representation of the different cases. Upper part = full length copy. Lower part = deleted copy. Grey bar = transposable element. Black bar = microhomologies. Percentages (X—Y): X = percentage observed among the 142 deletions of the Papilionis Tribe; Y = percentage observed among the 181 deletions of all Lemi
The microhomology size varies from 2 to 11 bp, with a mean value of 4 bp and a standard deviation of 2 bp (Fig. 6a). Most of them are 3 or 4 bp long. The base composition of the microhomologies is biased toward A/T (79 %—mean value for all categories of deletions BPEE, BPNN and BPEN). Compared to the base composition of the reference elements (65 % for A. thaliana and 69 % for M. truncatula), this suggests that most of the deletions occur between A/T rich sequences. This particularly true for the microhomologies longer than 8 bp, which are stretches of A and T (for example TATTAATTTA or TTATTAATTTA). Moreover, the frequencies of microhomology sequences of 3 and 4 bp, are not higher than the other words of 3 and 4 bp present in the reference sequence (estimation done using Compseq of EMBOSS Explorer, data not shown). Finally, as shown in Fig. 6b, there is no relationship between microhomology size and deletion size. For the most common microhomology sizes (3 and 4 bp), a large set of deletion sizes can be observed. For longer microhomology sizes (higher the 8 bp), deletions seem short. However, it not possible to conclude since very few internal deletions with long microhomologies are available.
Several copies are relatively short and present deletions of more than 1,500 bp. The BPs of these deletions are localized within a region of 300 bp on either side of the element. A dotmatcher (EMBOSS Explorer) aligning the full-length reference copies against themselves clearly shows the existence of repeated sequences at both extremities of Lemi that are longer than the Inverted Terminal Repeat (28 bp—Figure 2 of supplementary data). More precisely, the analysis of the base composition of these regions shows that these extremities are extremely A/T rich compared to other elements of the Tc1-mariner-IS630 SuperFamily, such as the mariner and Impala elements and, to a lesser extent, to Tc1 (Table 2).
Discussion and conclusion
Our results have revealed new several features of the Lemi elements and raised many questions about their evolution. First, the known distribution of this element is so far restricted to a small fraction of the Eudicots. Second, internal deletions do not occur at random, but always involved microhomologies exactly or near the breaking points, which confirms and reinforces previous findings. Third, some of these microhomologies could be due to a distortion of the base composition, particularly at the ends of the elements. Fourth, this could have an evolutionary impact on the dynamics of Lemi.
The apparent restriction of the distribution of Lemi to a few Eudicots is probably due to the lack of genomic data for many species of this Order. This makes it impossible to infer a relevant evolutionary dynamic of this element between species (horizontal and/or vertical transfer), and we can only suspect or suggest putative scenarios. For instance, the fact that Lemi are present only in Eudicots could be evidence in favor of vertical transfers. Indeed, based on the genomes available, the presence of large numbers of deleted copies prevents successful horizontal transfers (i.e. a transfer followed by multiplication in the genome of the naive species) since most of the copies are non-autonomous. This hypothesis seems to be confirmed within the papilionis Tribe. However, no generalization can be drawn from these data, because very few genomes are available in Eudicots. Moreover, the ploidization level of all species is not the same and no estimates of the age of the copies can be done without any information on the average mutation rate in Eudicots. Therefore, without an extensive analysis, it could be difficult to solve this question.
With regard to the second feature, several mechanisms of DNA repair could give rise to deletions associated with microhomologies and at or close to the BPs (McVey and Lee 2008; Puchta 2005; Cockram et al. 2007). The abortive gap repair (AGR) mechanism proposed by (Rubin and Levy 1997) is a mechanism based on synthesis-dependent strand annealing (SDSA). More precisely, after the excision of a Class II element, SDSA starts to fill the gap left, using another copy as template. However, this mechanism can abort before the end of the complete repair. This usually leads to the emergence of several categories of non-autonomous copies with an internal deletion due to the premature ending of repair followed by the annealing of the two neo-synthesized strands through microhomologies. This mechanism of repair can also lead to deleted copies with short insertions of sequence that do not belong to the element. This is interpreted as the result of a template switch during the gap repair. Finally, duplication of short internal sequences due to slippage can also be observed (Rubin and Levy 1997). In Drosophila a mechanism of this type has been proposed to explain the abundance of copies with internal deletion(s) and the existence of rare deleted copies with a short internal insertion (Brunet et al. 2002).
Other mechanisms can be also involved in such a phenomenon (McVey and Lee 2008; Puchta 2005). They include non-homologous end joining (NHEJ), microhomology end joining (MMEJ), and single strand annealing (SSA). All of them are able to generate small or large deletions due to annealing of microhomologies from less than 5 bp (NHEJ) up to more than 30 bp, as in SSA. Moreover, several authors have shown or suggested that TEs (like Sleeping Beauty or Ac/Ds) can interact with the protein involved in NHEJ mechanisms like Ku (Izsvak et al. 2004; Yant and Kay 2003; Yu et al. 2004). More recently, it was suggested “that blunt joins, junctional microhomologies and short indels (deletion with insertion)” … fit the model of SD-MMEJ [synthesis-dependent microhomology-mediated end joining (Yu and McVey 2010)]. This mechanism occurs in most eukaryotes, including plants, through the Mer11 complex (Heacock et al. 2004; Decottignies 2007).
These features have raised several questions about the evolutionary dynamics of Class II elements, and in particular of Lemi. Due to their mode of transposition (cut and paste), Class II TEs can lead to DSBs. Whatever the repair mechanism involved, most of them can produce internal deletion between microhomologies. This could be a quick way to reduce their activity, and their genome invasion and impact on the genesis of non-autonomous copies. Indeed, when TEs are naturally or artificially injected into a naive genome, genome invasion is followed by various different dynamics, depending on their rates of transposition, excision, and mutation, and their global activities (Le Rouzic et al. 2007). These dynamics are generally the result of competition between autonomous and non-autonomous copies. Indeed, it has already been reported that several deleted copies can regulate the activity of autonomous copies, such as the KP copies, and the activity of the full-length P (Brookfield 1996), or the deleted mariner copy found in D. teissieri on the mos1 copy (De Aguiar and Hartl 1999) and many short variants of Tol2 (Koga et al. 2011).
Another question that arises is whether the microhomology frequency higher in TEs than in genes, or is the frequency of internal deletion in TEs due solely to their mode of transposition leading to DSB. At present, it is still difficult to answer these questions because several parameters of TE dynamics are not known (amplification rate of non-autonomous copies vs. that of autonomous ones, relationship between the excision rate and the deletion rate…), preliminary results do not suggest a higher frequency of microhomologies in TEs than in genes (data not shown). Therefore, the rapid emergence of TE copies with internal deletion could be related the high frequency of transposition during the first step of genome invasions (Le Rouzic and Capy 2005).
Compared to other Class II elements, such as mariner, Impala or Osmar1, Lemi seems to have a set of features that are probably important for the emergence of copies with large internal deletions i.e. copies of less than 600 bp. To produce such copies in Lemi, deletions have to occur between sequences present at both extremities of the full-length copies. These regions (5′ and 3′) are particularly A/T rich (82 % for 5′ end vs. 76 % for 3′ end in A. thaliana and, 75 vs. 73 % in M. truncatula), thus increasing the probability of observing microhomologies involving stretches of A or T. For instance, the probability of observing the AAA motif when the A frequency is 50 % is equal to 0.125, and becomes equal to 0.016 when the A frequency is 25 %. However if such a phenomenon can enhance the emergence of MITEs, then their genomic amplification is probably due to other features. For instance, elements like Tc1 present an A/T richness in their extremities that is close to 70 %, and the genome of Caenorhabditis elegans contains about 2 % of MITEs (Bessereau 2006). However, in the Impala Family, the A/T richness of the extremities is close to 50 %, and MITES such as mimp have also been described (Dufresne et al. 2007). Similarly, the relationship between Stowavay and the Plant mariner-like element (Feschotte et al. 2003; Yang et al. 2009) does not seem to be due to the existence of high frequency of microhomologies in these elements.
If such characteristics can be responsible for the emergence of MITEs, other mechanisms can be suggested like the existence of a predominant DNA repair mechanism(s) or TE inactivation/recognition mechanism that could induce internal deletions. Moreover, the propagation of the deleted copies in a genome intuitively depends on the dynamics of active copies. If deletions occur during the invasion phase deleted copies will be amplified and a competition between autonomous and non-autonomous copies can settle (Le Rouzic et al. 2007), but if they occur during a period where the transposition rate is reduced, their copy number will probably remain relatively low.
In conclusion, analysis of deleted elements is of particular interest. Their origin (gap repair mechanisms or de novo), their own dynamics compared to those of autonomous copies, and their impact on the general activity of the Family help us to understand the short-term and long-term evolution of TEs, and their coevolution with their hosts. Therefore, all these informations will be useful to provide more realistic theoretical models of TE dynamics and evolution.
References
Achaz G, Coissac E, Viari A, Netter P (2000) Analysis of intrachromosomal duplications in yeast Saccharomyces cerevisiae: a possible model for their origin. Mol Biol Evol 17:1268–1275
Anxolabehere D, Benes H, Nouaud D, Periquet G (1987) Evolutionary steps and transposable elements in Drosophila melanogaster: the missing RP type obtained by genetic transformation. Evolution 41:846–853
Auge-Gouillou C, Hamelin MH, Demattei MV, Periquet M, Bigot Y (2001) The wild-type conformation of the Mos-1 inverted terminal repeats is suboptimal for transposition in bacteria. Mol Genet Genomics 265:51–57
Bessereau J-L (2006) Transposons in C. elegans. In: WormBook (ed.) The C. elegans research community. WormBook. doi:10.1895/wormbook.1.70.1, http://www.wormbook.org
Brookfield JFY (1996) Genetic evidence for repression of somatic P element movements in Drosophila melanogaster consistent with a role for the KP element. Heredity 76:384–391
Brunet F, Giraud T, Godin F, Capy P (2002) Do deletions of the Mos1-like elements occur randomly in the Drosophilidae family? J Mol Evol 54:227–234
Capy P, Anxolabehere D, Langin T (1994) The strange phylogenies of transposable elements: are horizontal transfers the only explantation? Trends Genet 10:7–12
Casacuberta E, Casacuberta JM, Puigdomenech P, Monfort A (1998) Presence of miniature inverted-repeat transposable elements (MITEs) in the genome of Arabidopsis thaliana: characterisation of the Emigrant family of elements. Plant J 16:79–85
Cockram J, Mackay IJ, O’Sullivan DM (2007) The role of double-stranded break repair in the creation of phenotypic diversity at cereal VRNI loci. Genetics 177:2535–2539
Daniels SB, Clark SH, Kidwell MG, Chovnick A (1987) Genetics transformation of Drosophila melanogaster with an autonomous P-element: phenotypic and molecular analyses of long-established transformed lines. Genetics 115:711–723
De Aguiar D, Hartl DL (1999) Regulatory potential of nonautonomous mariner elements and subfamily crosstalk. Genetica 107:79–85
Decottignies A (2007) Microhomology-Mediated end joining in fission yeast is repressed by Pku70 and relies on genes involved in homologous recombination. Genetics 176:1403–1415
Dufresne M, Hua-Van A, Abd el Wahab H, Ben M’Barek S, Kerma GHL, Daboussi MJ (2007) Transposition of a fungal MITE through the action of a Tc1-like transposase. Genetics 175:441–452
Feschotte C, Mouches C (2000) Evidence that a family of miniature inverted-repeat transposable elements (MITEs) from the Arabidopsis thaliana genome has arisen from a pogo-like DNA transposon. Mol Biol Evol 17:730–737
Feschotte C, Zhang X, Wessler SR (eds) (2002) Miniature inverted-repeat transposable elements and their relationship to Eetablished DNA transposons. Mobile DNA II. ASM Press, Washington, DC, USA
Feschotte C, Swamy L, Wessler SR (2003) Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with Stowaway miniature inverted repeat transposable elements (MITEs). Genetics 163:747–758
Galindo MI, Ladeveze V, Lemeunier F, Kalmes R, Periquet G, Pascual L (1995) Spread of the autonomous transposable element hobo in the genome of Drosophila melanogaster. Mol Biol Evol 12:723–734
Gao CH, Xiao ML, Ren XD, Hayward A, Yin JM, Wu LK, Fu DH, Li JN (2012) Characterization and functional annotation of nested transposable elements in eukaryotic genomes. Genomics 100:222–230
Grover CE, Kim HR, Wing RA, Paterson AH, Wendel JF (2004) Incongruent patterns of local and global genome size evolution in cotton. Genome Res 14:1474–1482
Guermonprez H, Loot C, Casacuberta JM (2008) Different strategies to persist: the pogo-like Lemi1 transposon produces miniature inverted-repeat transposable elements or typical defective elements in different plant genomes. Genetics 180:83–92
Heacock M, Spangler E, Riha K, Puizina J, Shippen DE (2004) Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J 23:2304–2313
Hua-Van A, Daviere JM, Kaper F, Langin T, Daboussi MJ (2000) Genome organization in Fusarium oxysporum: clusters of Class II transposons. Curr Genet 37:339–347
Izsvak Z, Stuwe EE, Fiedler D, Katzer A, Jeggo PA, Ivics Z (2004) Healing the wounds inflicted by Sleeping Beauty transposition by double-strand break repair in mammalian somatic cells. Mol Cell 13:279–290
Jacobson JW, Hartl DL (1985) Coupled instability of two X-linked genes in Drosophila mauritiana: germinal and somatic instability. Genetics 111:57–65
Jacobson JW, Medhora MM, Hartl DL (1986) Molecular structure of a somatically unstable transposable element in Drosophila. Proc Natl Acad Sci USA 83:8684–8688
Jiang N, Feschotte C, Zhang XY, Wessler SR (2004) Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs). Curr Opin Plant Biol 7:115–119
Koga A, Sasaki S, Naruse K, Shimada A, Sakaizumi M (2011) Occurrence of a short variant of the Tol2 transposable element in natural populations of the medaka fish. Genet Res 93:13–21
Ladeveze V, Galindo I, Chaminade N, Pascual L, Periquet G, Lemeunier F (1998) Transmission pattern of hobo transposable element in transgenic lines of Drosophila melanogaster. Genet Res 71:97–107
Le Rouzic A, Capy P (2005) The first steps of transposable elements invasion: parasitic strategy vs. genetic drift. Genetics 169:1033–1043
Le Rouzic A, Boutin TS, Capy P (2007) Long-term evolution of transposable elements. Proc Natl Acad Sci USA 104:19375–19380
McVey M, Lee SE (2008) MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet 24:529–538
Puchta H (2005) The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot 56:1–14
Rouault JD, Casse N, Chenais B, Hua-Van A, Filee J, Capy P (2009) Automatic classification within families of transposable elements: Application to the mariner Family. Gene 448:227–232
Rubin E, Levy AA (1997) Abortive gap repair: underlying mechanism for Ds element formation. Mol Cell Biol 17:6294–6302
SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274:765–768
Scavarda NJ, Hartl DL (1987) Germ line abnormalities in Drosophila simulans transformed with the transposable P-element. J Genet 66:1–15
Soltis DE, Soltis PS, Chase MW, Mort ME, Albach DC, Zanis M, Savolainen V, Hahn WH, Hoot SB, Fay MF, Axtell M, Swensen SM, Prince LM, Kress WJ, Nixon KC, Farris JS (2000) Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot J Linn Soc 133:381–461
The Angiosperm Phylogeny Group (2003) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc 141:346–399
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Yang GJ, Nagel DH, Feschotte C, Hancock CN, Wessler SR (2009) Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science 325(5946):1391–1394
Yant SR, Kay MA (2003) Nonhomologous-end-joining factors regulate DNA repair fidelity during Sleeping Beauty element transposition in mammalian cells. Mol Cell Biol 23(23):8505–8518
Yu AM, McVey M (2010) Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acid Res 38:5706–5717
Yu JH, Marshall K, Yamaguchi M, Haber JE, Weil CF (2004) Microhomology-dependent end joining and repair of transposon-induced DNA hairpins by host factors in Saccharomyces cerevisiae. Mol Cell Biol 24:1351–1364
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10709_2013_9736_MOESM1_ESM.pdf
Supplementary data - Figure 1: Phylogeny of Lemi copies belonging to the papilionis Tribe. The minimum size of the copies is 2,035 bp. The phylogeny was constructed using the Neighbor-Joining method (default parameters, bootstrap with 1,000 replications) of MEGA. (PDF 57 kb)
10709_2013_9736_MOESM2_ESM.pdf
Supplementary data – Figure 2: Dotplot of mariner and Lemi elements of M. truncaluta (MtLemi1) and A. thaliana (AtLemi1) against themselves using dotmatcher of EMBOSS. (PDF 464 kb)
Rights and permissions
About this article
Cite this article
Negoua, A., Rouault, JD., Chakir, M. et al. Internal deletions of transposable elements: the case of Lemi elements. Genetica 141, 369–379 (2013). https://doi.org/10.1007/s10709-013-9736-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-013-9736-3