Abstract
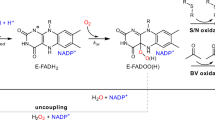
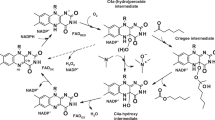
Flavin-containing monooxygenases (FMOs) metabolize xenobiotic compounds, many of which are clinically important, as well as endogenous substrates as part of a discrete physiological process. The FMO gene family is conserved and ancient with representatives present in all phyla so far examined. However, there is a lack of information regarding the long-term evolution and functional divergence of these proteins. This study represents the first attempt to characterize the long-term evolution followed by the members of this family. Our analysis shows that there is extensive silent divergence at the nucleotide level suggesting that this family has been subject to strong purifying selection at the protein level. Invertebrate FMOs have a polyphyletic origin. The functional divergence of FMOs 1–5 started before the split between amphibians and mammals. The vertebrate FMO5 is more ancestral than other four FMOs. Moreover, the existence of higher levels of codon bias was detected at the N-terminal ends, which can be ascribed to the critical role played by the FAD binding motif in this region. Finally, critical amino acid residues for FMO functional divergence (type I & II) after gene duplication were detected and characterized.







Similar content being viewed by others
References
Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105
Agosin M, Ankley GT (1987) Conversion of N, N-dimethylaniline to N, N-dimethylaniline-N-oxide by a cytosolic flavin-containing enzyme from Trypanosoma cruzi. Drug Metab Dispos 15:200–203
Alfieri A, Malito E, Orru R et al (2008) Revealing the moonlighting role of NADP in the structure of a flavin-containing monooxygenase. Proc Natl Acad Sci U S A 105:6572–6577
Allerston CK, Shimizu M, Fujieda M et al (2007) Molecular evolution and balancing selection in the flavin-containing monooxygenase 3 gene (FMO3). Pharmacogenet Genomics 17:827–839
Boulton CA, Crabbe MJ, Large PJ (1974) Microbial oxidation of amines. Partial purification of a trimethylamine mono-oxygenase from Pseudomonas aminovorans and its role in growth on trimethylamine. Biochem J 140:253–263
Cashman JR, Zhang J (2006) Human flavin-containing monooxygenases. Annu Rev Pharmacol Toxicol 46:65–100
Cherrington NJ, Cao Y, Cherrington JW et al (1998) Physiological factors affecting protein expression of flavin-containing monooxygenases 1, 3 and 5. Xenobiotica 28:673–682
da Fonseca RR, Antunes A, Melo A et al (2007) Structural divergence and adaptive evolution in mammalian cytochromes P450 2C. Gene 387:58–66
Eirín-López JM, Frehlick LJ, Ausió J (2006) Long-term evolution and functional diversification in the members of the nucleophosmin/nucleoplasmin family of nuclear chaperones. Genetics 173:1835–1850
Eswaramoorthy S, Bonanno JB, Burley SK et al (2006) Mechanism of action of a flavin-containing monooxygenase. Proc Natl Acad Sci U S A 103:9832–9837
Force A, Lynch M, Pickett FB et al (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545
Force A, Shashikant C, Stadler P et al (2004) Comparative genomics, cis-regulatory elements, and gene duplication. Methods Cell Biol 77:545–561
Glenn KL, Ramos AM, Rothschild MF (2007) Analysis of FMO genes and off flavour in pork. J Anim Breed Genet 124:35–38
Goldstone JV, Goldstone HM, Morrison AM et al (2007) Cytochrome P450 1 genes in early deuterostomes (tunicates and sea urchins) and vertebrates (chicken and frog): origin and diversification of the CYP1 gene family. Mol Biol Evol 24:2619–2631
Gu X (2006) A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol Biol Evol 23:1937–1945
Gu X, Vander Velden K (2002) DIVERGE: phylogeny-based analysis for functional-structural divergence of a protein family. Bioinformatics 18:500–501
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hao DC, Sun J, Furnes B et al (2006) Haplotype frequency distribution and linkage disequilibrium analysis of single nucleotide polymorphisms at the human FMO3 gene locus. Biochem Genet 44:391–407
Hao DC, Sun J, Furnes B et al (2007) Allele and genotype frequencies of polymorphic FMO3 gene in two genetically distinct populations. Cell Biochem Funct 25:443–453
Hernandez D, Janmohamed A, Chandan P et al (2004) Organization and evolution of the flavin-containing monooxygenase genes of human and mouse: identification of novel gene and pseudogene clusters. Pharmacogenetics 14:117–130
Hines RN, Cashman JR, Philpot RM et al (1994) The mammalian flavin-containing monooxygenases: molecular characterization and regulation of expression. Toxicol Appl Pharmacol 125:1–6
Honkatukia M, Reese K, Preisinger R et al (2005) Fishy taint in chicken eggs is associated with a substitution within a conserved motif of the FMO3 gene. Genomics 86:225–232
Kim YM, Ziegler DM (2000) Size limits of thiocarbamides accepted as substrates by human flavin-containing monooxygenase 1. Drug Metab Dispos 28:1003–1006
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Krueger SK, Williams DE (2005) Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther 106:357–387
Lattard V, Zhang J, Tran Q et al (2003) Two new polymorphisms of the FMO3 gene in Caucasian and African-American populations: comparative genetic and functional studies. Drug Metab Dispos 31:854–860
Lattard V, Zhang J, Cashman JR (2004) Alternative processing events in human FMO genes. Mol Pharmacol 65:1517–1525
Lunden A, Marklund S, Gustafsson V et al (2002) A nonsense mutation in the FMO3 gene underlies fishy off-flavor in cow’s milk. Genome Res 12:1885–1888
Lynch M, O’Hely M, Walsh B et al (2001) The probability of preservation of a newly arisen gene duplicate. Genetics 159:1789–1804
Mitchell SC, Smith RL (2001) Trimethylaminuria: the fish malodor syndrome. Drug Metab Dispos 29:517–521
Naumann C, Hartmann T, Ober D (2002) Evolutionary recruitment of a flavin-dependent monooxygenase for the detoxification of host plant-acquired pyrrolizidine alkaloids in the alkaloid-defended arctiid moth Tyria jacobaeae. Proc Natl Acad Sci U S A 99:6085–6090
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152
Ohmi N, Yoshida H, Endo H et al (2003) S-oxidation of S-methyl-esonarimod by flavin-containing monooxygenases in human liver microsomes. Xenobiotica 33:1221–1231
Overby LH, Buckpitt AR, Lawton MP et al (1995) Characterization of flavin-containing monooxygenase 5 (FMO5) cloned from human and guinea pig: evidence that the unique catalytic properties of FMO5 are not confined to the rabbit ortholog. Arch Biochem Biophys 317:275–284
Park CS, Kang JH, Chung WG et al (2002) Ethnic differences in allelic frequency of two flavin-containing monooxygenase 3 (FMO3) polymorphisms: linkage and effects on in vivo and in vitro FMO activities. Pharmacogenetics 12:77–80
Posada D (2006) ModelTest server: a web-based tool for the statistical selection of models of nucleotide substitution online. Nucleic Acids Res 34(Web Server issue):W700–W703
Rodríguez-Fuentes G, Aparicio-Fabre R, Li Q et al (2008) Osmotic regulation of a novel flavin-containing monooxygenase in primary cultured cells from rainbow trout (Oncorhynchus mykiss). Drug Metab Dispos 36:1212–1217
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rooney AP (2003) Selection for highly biased amino acid frequency in the TolA cell envelope protein of proteobacteria. J Mol Evol 57:731–736
Rooney AP, Zhang J, Nei M (2000) An unusual form of purifying selection in a sperm protein. Mol Biol Evol 2000(17):278–283
Rozas J, Sanchez-Del Barrio JC, Messeguer X et al (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schlenk D (1993) A comparison of endogenous and exogenous substrates of the flavin-containing monooxygenases in aquatic organisms. Aquat Toxicol 26:157–162
Schlenk D, Buhler DR (1989) Xenobiotic biotransformation in the Pacific oyster (Crassostrea gigas). Comp Biochem Physiol C 94:469–475
Schlenk D, Yeung C, Rettie A (2004) Unique monooxygenation pattern indicates novel flavin-containing monooxygenase in liver of rainbow trout. Mar Environ Res 58:499–503
Suh JK, Poulsen LL, Ziegler DM et al (1999) Yeast flavin-containing monooxygenase generates oxidizing equivalents that control protein folding in the endoplasmic reticulum. Proc Natl Acad Sci U S A 96:2687–2691
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland
Tamura K, Dudley J, Nei M et al (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thomas JH (2007) Rapid birth-death evolution specific to xenobiotic cytochrome P450 genes in vertebrates. PLoS Genet 3:e67
Veeramah KR, Thomas MG, Weale ME et al (2008) The potentially deleterious functional variant flavin-containing monooxygenase 2*1 is at high frequency throughout sub-Saharan Africa. Pharmacogenet Genomics 18:877–886
Wapinski I, Pfeffer A, Friedman N et al (2007) Natural history and evolutionary principles of gene duplication in fungi. Nature 449:54–61
Wen Z, Rupasinghe S, Niu G et al (2006) CYP6B1 and CYP6B3 of the black swallowtail (Papilio polyxenes): adaptive evolution through subfunctionalization. Mol Biol Evol 23:2434–2443
Wright F (1990) The ‘effective number of codons’ used in a gene. Gene 87:23–29
Yeung CK, Adman ET, Rettie AE (2007) Functional characterization of genetic variants of human FMO3 associated with trimethylaminuria. Arch Biochem Biophys 464:251–259
Zhang J, Cashman JR (2006) Quantitative analysis of FMO gene mRNA levels in human tissues. Drug Metab Dispos 34:19–26
Zhang M, Robertus JD (2002) Molecular cloning and characterization of a full-length flavin-dependent monooxygenase from yeast. Arch Biochem Biophys 403:277–283
Zhang J, Rosenberg HF, Nei M (1998) Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci USA 95:3708–3713
Zschocke J, Kohlmueller D, Quak E et al (1999) Mild trimethylaminuria caused by common variants in FMO3 gene. Lancet 354:834–835
Zwickl DJ (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation, The University of Texas at Austin
Acknowledgments
This study was supported by China Postdoctoral Science Foundation (project 20080440019), Education Department of Liaoning Province (2009A120), and the Start-up Research Fund (2008) of Dalian Jiaotong University of China. We are grateful to Dr. William J. Etges and the anonymous reviewers for their critical comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Hao, D.C., Chen, S.L., Mu, J. et al. Molecular phylogeny, long-term evolution, and functional divergence of flavin-containing monooxygenases. Genetica 137, 173–187 (2009). https://doi.org/10.1007/s10709-009-9382-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-009-9382-y