Abstract
Post mortem storage is a necessary process for removal of pin bones without destruction of fillets, thereby avoiding volume and economic loss. However, the enzymes involved in loosening pin bones during storage have not been studied to a great extent. In this study, the activities and localization of MMPs in the connective tissue (CT) of pin bones dissected from fillet of salmon and cod were investigated. Interestingly, the enzyme activity profile in these two species was different during post mortem storage of fish fillets. Adding MMP inhibitor (GM6001) and serine protease inhibitor (Pefabloc) revealed different effects in the two species, suggesting different regulations in salmon and cod. In situ zymography with the same inhibitors verified MMP and serine protease activity in CT close to pin bone at early post mortem (6 h) in salmon. However, MMP inhibition was not evident in cod in this area at that time point. Immunohistochemistry further revealed MMP9 and MMP13 were located more to the outer rim of CT, facing the pin bone and adipose tissue, while MMP7 was more randomly distributed within CT in salmon. In contrast, all these three MMPs were randomly distributed in CT in cod. In summary, our study reveals different MMP enzyme profiles in salmon and cod in the pin bone area, influenced by serine proteases, and suggests that MMPs and serine proteases must be taken in consideration when studying the conditions for early pin bone removal.
Similar content being viewed by others
Introduction
The consumers prefer fresh and boneless fish fillets. However, a major problem associated with pin bones is the firm attachment to the muscles which can lead to fillet destruction during removal or breakage halfway into the fillet, so that only a portion of the pin bones is removed. The fish industry needs knowledge about optimal conditions for pin bone removal to save time and costs. So far, little is known about the enzymes responsible for weakening the connective tissue (CT) surrounding the pin bones and the attachment to the surrounding tissue. There are major differences between salmon and cod in terms of bone strength and pulling force required to remove the pin bones (Akse and Tobiassen 2002; Esaiassen and Sørensen 1996; Westavik 2009). Whether this is due to a specific difference in enzymatic profiles post mortem in the two species is currently unknown. Degradation of the CT is enzymatic, involving numerous enzymes that can be regulated by various factors including pH, temperature and ion strength and processes that affect these factors could as such impact loosening of the pin bones (Larsen et al. 2008; Vargova et al. 2012).
Proteases are central for CT degradation and are grouped based on their catalytic residues, matrix metalloproteases (MMPs), serine proteases, cysteine proteases, threonine proteases and aspartic proteases (Cawston and Wilson 2010). MMPs are the major group of proteases important for extracellular matrix (ECM) degradation. They are classified based on their substrate specificities and include collagenases (MMPs 1, 8, 13), gelatinases (MMPs 2, 9), matrilysins (MMPs 7, 11, 26) and stromelysins (MMPs 3, 10) (see (Pedersen et al. 2015) for review of MMPs in fish). The MMPs are normally secreted as zymogens, which are subsequently processed by proteolytic enzymes to generate the active forms (Okumura et al. 1997; Woessner 1991). Under normal physiological conditions, the proteolytic activity of the MMPs is controlled at any of the following three known stages: transcription, activation of the zymogens and inhibition of the active forms by various tissue inhibitors of MMPs (TIMPs) (Verma and Hansch 2007). Extracellular proteases influence and activate each other in a complex network, and often one protease pathway is combined with another (He et al. 1989; Shamamian et al. 2001; Zhu et al. 2001).
In this study, we compared extracellular enzymes present in the attachment area of pin bones in salmon and cod during the post mortem period. The aim was to investigate the specific distribution of MMP activities in this specific area. Samples were harvested at different time points post mortem, and enzyme expression and activities around pin bones area were investigated by immunohistochemistry and in situ zymography. Our results reveal new and important distribution patterns of MMPs in salmon and cod and also differences between the two species during post mortem storage.
Materials and methods
Fish samples
Tissues were obtained from salmon (Salmo salar) and cod (Gadus morhua L.). Fillets harvested immediately after slaughter were stored on ice for 0 min, 6, 12, 24 h or 5 days. For total MMP activity assay, pin bones with surrounding CT (including some residues of surrounding adipose/muscle tissue) were dissected, snap frozen in liquid nitrogen and stored at −80 °C until further analysis. For microscopy studies, pieces including pin bone area of approximately 15 × 10 × 10 mm were cut from anterior positions in the fish fillets and fixed in zinc-buffered fixative (36.7 mM ZnCl2, 27.3 mM ZnAc2 × 2H2O, 0.63 mM CaAc2 in 0.1 M Tris, pH 7.4) for 36–38 h. Thereafter, the samples were decalcified with EDTA (14 %, pH 7.1 at RT) for 10 days, before dehydration and paraffin embedding.
Protein extraction from pin bone tissue
The frozen pin bone tissue from salmon or cod were homogenized in 2-ml tubes with 2.8-mm ceramic beads (Precellys) and 20 µl of lysis buffer (0.25 % Triton X-100 in 10 mM CaCl2 and 100 mM HEPES, pH 7.0) per mg tissue using Precellys 24 tissue homogenizer at 5.000 rpm for 4 × 30 s. The tubes were then incubated on ice for 1 h before being centrifuged at 13.000 rpm for 15 min at 4 °C. Protein concentration in the supernatants was determined with BCA protein assays (Thermo Fisher Scientific) before storage at −80 °C until further analysis.
MMP activity assay
Total MMP activity was measured using SensoLyte 520 generic MMP assay kit fluorometric (AnaSpec). In brief, equal amount (3 µg) of pin bone tissue extracts was added to a 96-well plate and diluted with assay buffer to 50 µl/well. For inhibition assay, 0.5 mM GM6001 (Calbiochem) or 8 mM Pefabloc (Roche) was added to the samples. A blank control of only assay buffer was also included. A total of 50 µl/well of the generic MMP substrate solution was added to the sample and control wells, and the reagents were mixed by gentle shaking the plate for 30 s. The plate was incubated at RT for 1 h in darkness before the fluorescence intensity was measured at Ex/Em = 490/520 nm with a fluorometric microplate reader. The MMP activity was expressed as relative fluorescence unit (RFU) by subtracting the background reading from the sample readings.
In situ zymography
The method was performed as described in Hadler-Olsen et al. (2010). In brief, 5-µm-thick sections from ZBF-fixed and paraffin-embedded pin bone tissue (6 h) were heated at 58 °C overnight, deparaffinized in xylene and rehydrated in graded alcohol baths. Two hundred milliliters substrate of dye-quenched (DQ) gelatin (Invitrogen), DQ-collagen or DQ-casein (Life Technologies) diluted 1:50 in reaction buffer (50 mM Tris–HCl, 150 mM NaCl, 5 mM CaCl2, 0.2 mM sodium azide, and pH 7.6) was added to the tissue sections and incubated in dark humidity chamber at 37 °C for 2 h. To evaluate the contribution of proteases, sections were pre-incubated with 0.5 mM GM6001 or 8 mM Pefabloc for 1 h at 37 °C. Sections were then rinsed 2 × 5 min in PBS baths, dipped in Milli-Q water and air-dried for few minutes. The sections were mounted using SlowFade Gold antifade reagent with DAPI (Invitrogen) and examined with a confocal microscope Olympus FluoView FV1000 (Olympus).
Immunohistochemistry
Sections were deparaffinized and rehydrated before permeabilizing with 0.5 % Triton-X100 in PBS for 15 min. Non-specific antibody binding was blocked by incubating the sections with 5 % BSA for 1 h. The sections were incubated with mouse anti-MMP7 (Santa Cruz), rabbit anti-MMP9 (Novus Biological) or rabbit anti-MMP13 (Abcam) antibodies (all diluted 1:50) for 2 h, before washing with PBS and subsequently incubation with Alexa 546-conjugated secondary antibodies (Invitrogen, 1:600) for 45 min. Sections were washed in PBS, mounted with SlowFade Gold antifade reagent with DAPI and examined with a confocal microscope Olympus FluoView FV1000. The images were processed using Adobe Photoshop CS4 (Adobe Systems Inc.), brightness and contrast, if used were adjusted manually across the entire image.
Results and discussion
Our results show that the total MMP activity in salmon increased significantly short time after slaughter (6 h), followed by a dramatic reduction already after 12 h storage (Fig. 1a, left panel). This is in contrast to the MMP enzyme profile in cod, where the activities of MMPs sustained during storage, with just a minor increase at 12 and 24 h (Fig. 1a, right panel). To characterize the regulation of MMP activity in the two species, we added an MMP inhibitor (GM6001) and a serine protease inhibitor (Pefabloc) to the samples collected at 6 h before and we measured the MMP activity (Fig. 1b). Interestingly, our experiments demonstrate that the MMP activity was regulated differently in salmon and cod. Our results show that adding GM6001 inhibited total MMP activity in both salmon and cod. However, when adding Pefabloc, the total MMP activity was inhibited in salmon, while no effect on activity was observed in cod. This suggests that serine proteases are important for MMP activation in salmon, but not to the same degree in cod. Our unpublished proteomic data analysis (Rønning et al. 2016 under revision by Fish Physiology and Biochemistry) also shows a different protein composition and expression in salmon and cod. In addition, there are changes in a relatively large number of proteins during storage, indicating that the connective tissue surrounding pin bone is subject to multiple post mortem changes. Differences in expression of enzymes and regulatory proteins could possibly result in different regulations and kinetics in the two species.
Total MMP activity in pin bone area of salmon and cod determined using generic MMP activity assay. a MMP activity during post mortem storage on ice (n = 10). b MMP activity measured in presence of the universal MMP inhibitor GM6001 and serine protease inhibitor Pefabloc (n = 4). The results are expressed as mean ± SEM. *p ≤ 0.05 and **p ≤ 0.01 indicate statistically significant difference between the 0 h and the storage times post mortem, or between untreated and inhibitor treatments, as evaluated by the Student’s unpaired t test
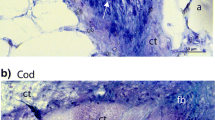
In situ zymography visualizes the precise localization of the enzyme activities in the tissue. Our experiment with MMP substrate DQ-gelatin demonstrated the presence of MMP activity in the CT surrounding pin bones and in the surrounding muscle and adipose tissue of salmon (Fig. 2, left panel). Using GM6001 and Pefabloc, the gelatinolytic activity in the CT was inhibited (Fig. 2, middle and right panel). Using different substrates, (gelatin, collagen and casein) we demonstrated MMP activity and serine protease activity in the CT close to the pin bones, summarized in Table 1. Inhibition of the enzyme activity in the CT close to pin bone was less visible in cod (Table 1), most likely reflecting less MMP activity present at the time point studied (6 h). Interestingly, although the enzyme activity was clearly inhibited in the CT, the activity in the surrounding tissue was not depressed by MMP or serine protease inhibitors, revealing a different enzyme profile in the CT close to pin bone compared to the CT in surrounding skeletal muscle and adipose tissue. MMPs exhibit a broad range of substrate specificities, including ECM proteins as well as non-ECM proteins. Collagen and gelatin are preferred substrates for the collagenase family and gelatinase family, respectively, although they can also be cleaved by other MMPs (Nagase 2001). Casein is a less common and preferred substrate for MMPs, but are frequently used in zymography for determining activity of MMP1 and MMP7 (Hu and Beeton 2010; Snoek-van Beurden and Von den Hoff 2005; Zeng et al. 2002). Casein is also a substrate for serine proteases. Under normal physiological processes, MMPs must be expressed to the exact extra- or peri-cellular location, at the right time and in the right amount. Also, they must be activated or inhibited appropriately. Most MMPs are synthesized and secreted as inactive proenzymes, and plasmin has been described as a key activator of several MMPs (He et al. 1989; Murphy et al. 1999). However, many other serine proteases have also been shown to directly activate MMPs in vitro or in vivo (Duncan et al. 1998; Fang et al. 1997; Gruber et al. 1989; Okada et al. 1987; Shamamian et al. 2001). In all cases, the activation requires the disruption of the Cys-Zn2+ interaction in their active center, and the removal of the propeptide that often proceeds in a stepwise manner involving the actions both from serine proteases and from activated MMPs (Gruber et al. 1989; Shamamian et al. 2001; Zhu et al. 2001).
In situ zymography of salmon pin bone tissue section with DQ-gelatin in presence or absence of the universal MMP inhibitor GM6001 and the serine protease Pefabloc. Arrows point out the CT surrounding the pin bone. Pb pin bone; CT connective tissue; A adipose tissue; MT muscle tissue. Scale bars as indicated
Using immunofluorescence, we demonstrate the presence of MMP7, MMP9 and MMP13 in the CT close to the pin bones (Fig. 3). In salmon, MMP9 and MMP13 were located to the outer rim of the CT, facing the pin bone and adipose tissue, while MMP7 was distributed more randomly. This is in contrast to the location in cod, where the MMPs were randomly distributed in the CT surrounding pin bones. Experiments in our laboratory have shown that the degradation of the CT in salmon and cod is different during post mortem storage (unpublished data). In salmon, the loosening occurred at the interface of pin bone and CT during degradation. This is in contrast to a more even degradation within the CT of cod. The different MMP distribution patterns in salmon and cod, especially MMP9 and MMP13, should be of interest for further studies. Proteolysis often occurred in the immediate vicinity of the cell in peri-cellular pockets close to the cell membrane where MMPs can be secreted to specific areas at the cell surface. Such localization mechanisms could possibly allow a high degree of control and can enhance MMP activity, prevent access of MMP inhibitors, concentrate MMPs to their precise target substrates and limit the extent of proteolysis to a defined region (Zucker et al. 2003).
There are several MMPs identified in fish, but the precise functions of these are not well characterized yet (see (Pedersen et al. 2015) for review). MMP9 and MMP13 have been detected at mRNA levels in salmon bone tissue (Ytteborg et al. 2010). In bone, the removal of the outer osteoid layer by MMPs precedes the attachment of the osteoclast and the subsequent breakdown of the ECM by cysteine proteinases (Everts et al. 1992). MMPs have been proposed to participate in degradation of fish muscle during storage (Kubota et al. 2001, 2003; Lodemel et al. 2004; Wu et al. 2008), and both MMP2 and MMP9 have been demonstrated in muscle fillet of Atlantic cod, spotted wolfish and Atlantic salmon (Lødemel and Olsen 2003). In common carp, MMP2 plays a critical role in muscle softening by degradation of type I and V collagens (Xu et al. 2015). By immunohistochemistry, we could observe expression of MMP2 and MMP9 in connective tissue of skeletal muscle in our salmon and cod samples (data not shown). However, only MMP9 was detectable in the pin bone tissue (Fig. 3), suggesting that MMP2 is not a major contributor in softening of connective tissue surrounding pin bones. Enzyme analysis of firm and soft fillets from Atlantic salmon revealed more active MMPs in the softer muscles (Martinez et al. 2011). In addition, the presence of serine protease activity in skeletal muscle of red sea bream and hake has been demonstrated and suggested to be involved in texture tenderization of the fish muscle (Martone et al. 1991; Wu et al. 2010).
Conclusion
In this study, we have compared extracellular enzymes present in the attachment area of pin bones in salmon and cod during the post mortem periods. Our results demonstrate that salmon and cod have a different enzyme profile, with a different distribution of MMPs in the CT. Further, we show that there is a complex network of MMPs and serine proteases influencing each other, making both MMPS and serine proteases interesting targets for further studies optimizing the early pin bone removal process in salmon industry.
References
Akse L, Tobiassen T. (2002) Tykkfiskbein i torskefilet vol RApport 15/2002. Fiskeriforskning, AS
Cawston TE, Wilson AJ (2010) Understanding the role of tissue degrading enzymes and their inhibitors in development and disease. Best Pract Res Clin Rheumatol 20:983–1002. doi:10.1016/j.berh.2006.06.007
Duncan ME, Richardson JP, Murray GI, Melvin WT, Fothergill JE (1998) Human matrix metalloproteinase-9: activation by limited trypsin treatment and generation of monoclonal antibodies specific for the activated form. Eur J Biochem 258:37–43
Esaiassen M, Sørensen NK (1996) Fjerning av tykkfiskbein i laks vol Rapport 28/1996. Fiskeriforskning, AS
Everts V, Delaisse JM, Korper W, Niehof A, Vaes G, Beertsen W (1992) Degradation of collagen in the bone-resorbing compartment underlying the osteoclast involves both cysteine-proteinases and matrix metalloproteinases. J Cell Physiol 150:221–231. doi:10.1002/jcp.1041500202
Fang KC, Raymond WW, Blount JL, Caughey GH (1997) Dog mast cell alpha-chymase activates progelatinase B by cleaving the Phe88-Gln89 and Phe91-Glu92 bonds of the catalytic domain. J Biol Chem 272:25628–25635
Gruber BL, Marchese MJ, Suzuki K, Schwartz LB, Okada Y, Nagase H, Ramamurthy NS (1989) Synovial procollagenase activation by human mast cell tryptase dependence upon matrix metalloproteinase 3 activation. J Clin Investig 84:1657–1662. doi:10.1172/jci114344
Hadler-Olsen E, Kanapathippillai P, Berg E, Svineng G, Winberg JO, Uhlin-Hansen L (2010) Gelatin in situ zymography on fixed, paraffin-embedded tissue: zinc and ethanol fixation preserve enzyme activity. J Histochem Cytochem 58:29–39. doi:10.1369/jhc.2009.954354
He CS, Wilhelm SM, Pentland AP, Marmer BL, Grant GA, Eisen AZ, Goldberg GI (1989) Tissue cooperation in a proteolytic cascade activating human interstitial collagenase. Proc Natl Acad Sci U S A 86:2632–2636
Hu X, Beeton C (2010) Detection of functional matrix metalloproteinases by zymography. J Vis Exp. doi:10.3791/2445
Kubota M, Kinoshita M, Kubota S, Yamashita M, Toyohara H, Sakaguchi M (2001) Possible implication of metalloproteinases in post-mortem tenderization of fish muscle. Fish Sci 67:965–968. doi:10.1046/j.1444-2906.2001.00347.x
Kubota M, Kinoshita M, Takeuchi K, Kubota S, Toyohara H, Sakaguchi M (2003) Solubilization of type I collagen from fish muscle connective tissue by matrix metalloproteinase-9 at chilled temperature. Fish Sci 69:1053–1059. doi:10.1046/j.1444-2906.2003.00726.x
Larsen R, Olsen SH, Kristoffersen S, Elvevoll EO (2008) Low salt brining of pre-rigor filleted farmed cod (Gadus morhua L.) and the effects on different quality parameters Lwt-Food. Sci Technol 41:1167–1172. doi:10.1016/j.lwt.2007.07.015
Lødemel JB, Olsen RL (2003) Gelatinolytic activities in muscle of Atlantic cod (Gadus morhua), spotted wolffish (Anarhichas minor) and Atlantic salmon (Salmo salar). J Sci Food Agric 83:1031–1036. doi:10.1002/jsfa.1501
Lodemel JB, Maehre HK, Winberg JO, Olsen RL (2004) Tissue distribution, inhibition and activation of gelatinolytic activities in Atlantic cod (Gadus morhua). Comp Biochem Physiol B Biochem Mol Biol 137:363–371. doi:10.1016/j.cbpc.2003.12.007
Martinez I et al (2011) Protein expression and enzymatic activities in normal and soft textured Atlantic salmon (Salmo salar) muscle. Food Chem 126:140–148. doi:10.1016/j.foodchem.2010.10.090
Martone CB, Busconi L, Folco EJ, Sanchez JJ (1991) Detection of a trypsin-like serine protease and its endogenous inhibitor in hake skeletal muscle. Arch Biochem Biophys 289:1–5
Murphy G, Stanton H, Cowell S, Butler G, Knauper V, Atkinson S, Gavrilovic J (1999) Mechanisms for pro matrix metalloproteinase activation. APMIS 107:38–44
Nagase H (2001) Substrate Specificity of MMPs. In: Clendeninn NJ, Appelt K (eds) Matrix metalloproteinase inhibitors in cancer therapy. Humana Press, Totowa, pp 39–66. doi:10.1007/978-1-59259-011-7_2
Okada Y, Nagase H, Harris ED Jr (1987) Matrix metalloproteinases 1, 2, and 3 from rheumatoid synovial cells are sufficient to destroy joints. J Rheumatol 14(Spec No):41–42
Okumura Y, Sato H, Seiki M, Kido H (1997) Proteolytic activation of the precursor of membrane type 1 matrix metalloproteinase by human plasmin: A possible cell surface activator. FEBS Lett 402:181–184
Pedersen ME, Vuong TT, Ronning SB, Kolset SO (2015) Matrix metalloproteinases in fish biology and matrix turnover. Matrix Biol 44–46:86–93. doi:10.1016/j.matbio.2015.01.009
Rønning SB, Østbye TK, Krasnov A, Vuong TT, Veiseth-Kent E, Kolset SO, Pedersen ME (2016) A study of the firm attachment of pin bones in farmed salmon (Salmo salar) and cod (Gadus morhua L.) – the role of extracellular matrix components. Under revsion by Fish Physiology and Biochemistry
Shamamian P, Schwartz JD, Pocock BJ, Monea S, Whiting D, Marcus SG, Mignatti P (2001) Activation of progelatinase A (MMP-2) by neutrophil elastase, cathepsin G, and proteinase-3: a role for inflammatory cells in tumor invasion and angiogenesis. J Cell Physiol 189:197–206. doi:10.1002/jcp.10014
Snoek-van Beurden PA, Von den Hoff JW (2005) Zymographic techniques for the analysis of matrix metalloproteinases and their inhibitors. Biotechniques 38:73–83
Vargova V, Pytliak M, Mechirova V (2012) Matrix metalloproteinases. EXS. 103:1–33. doi:10.1007/978-3-0348-0364-9_1
Verma RP, Hansch C (2007) Matrix metalloproteinases (MMPs): chemical-biological functions and (Q)SARs. Bioorg Med Chem 15:2223–2268. doi:10.1016/j.bmc.2007.01.011
Westavik H (2009) Fjerning av pinnebein i filet av laks slaktet ved oppdrettsmerd. SINTEF Fiskeri og Havbruk, AS
Woessner JF Jr (1991) Matrix metalloproteinases and their inhibitors in connective tissue remodeling. FASEB J 5:2145–2154
Wu J-L, Lu B-J, Du M-H, Liu G-M, Hara K-J, Su W-J, Cao M-J (2008) Purification and characterization of gelatinase-like proteinases from the dark muscle of common carp (Cyprinus carpio). J Agric Food Chem 56:2216–2222. doi:10.1021/jf0728808
Wu GP, Chen SH, Liu GM, Yoshida A, Zhang LJ, Su WJ, Cao MJ (2010) Purification and characterization of a collagenolytic serine proteinase from the skeletal muscle of red sea bream (Pagrus major). Comp Biochem Physiol B: Biochem Mol Biol 155:281–287. doi:10.1016/j.cbpb.2009.11.014
Xu C et al (2015) Matrix Metalloproteinase 2 (MMP-2) Plays a critical role in the softening of common carp muscle during chilled storage by degradation of type I and V collagens. J Agric Food Chem 63:10948–10956. doi:10.1021/acs.jafc.5b03893
Ytteborg E, Torgersen J, Baeverfjord G, Takle H (2010) Morphological and molecular characterization of developing vertebral fusions using a teleost model. BMC Physiol 10:13
Zeng ZS, Shu WP, Cohen AM, Guillem JG (2002) Matrix metalloproteinase-7 expression in colorectal cancer liver metastases: evidence for involvement of MMP-7 activation in human cancer metastases. Clin Cancer Res 8:144–148
Zhu YK et al (2001) Synergistic neutrophil elastase-cytokine interaction degrades collagen in three-dimensional culture. Am J Physiol Lung Cell Mol Physiol 281:L868–L878
Zucker S, Pei D, Cao J, Lopez-Otin C (2003) Membrane type-matrix metalloproteinases (MT-MMP). Curr Top Dev Biol 54:1–74
Acknowledgments
This work was supported by grants from the Norwegian Seafood Research Fund FHF, Grant no. 900872.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Vuong, T.T., Rønning, S.B., Kolset, S.O. et al. The enzyme profiles in the connective tissue attaching pin bones to the surrounding tissue is specific in farmed salmon (Salmo salar) and cod (Gadus morhua L.). Fish Physiol Biochem 43, 19–25 (2017). https://doi.org/10.1007/s10695-016-0264-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-016-0264-9