Abstract
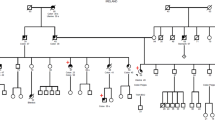
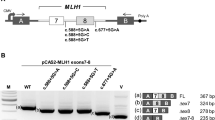
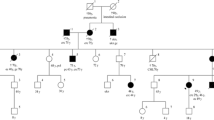
Lynch syndrome, also known as hereditary non-polyposis colorectal cancer, characterized by predisposition to colorectal cancer and other associated cancers, is an autosomal-dominant disorder mainly caused by germline mutations in DNA mismatch repair (MMR) genes such as MLH1, MSH2, and MSH6. Some mutations that disrupt splice donor or acceptor sites cause aberrant mRNA splicing. These mutations are generally considered as pathogenic ones, however, it is sometimes uneasy to accurately predict their pathogenicity without functional assays, particularly when the mutation is a single nucleotide substitution. In this report, we describe a 25-year-old patient with Lynch syndrome who carries a germline variant in a splice donor site of the MLH1 gene (c.790 + 5 G > T), which was first detected among Asian populations. The immunohistochemical analysis revealed loss of MLH1 protein expression in the tumor. Our splicing assay confirmed that the intronic MLH1 variant actually caused aberrant splicing, supporting its pathogenic effect. Our data accumulate more information on the genotype-phenotype relationships in patients with Lynch syndrome.



Similar content being viewed by others
References
Vasen HF, Moslein G, Alonso A, Bernstein I, Bertario L, Blanco I, Burn J, Capella G, Engel C, Frayling I, Friedl W, Hes FJ, Hodgson S, Mecklin JP, Moller P, Nagengast F, Parc Y, Renkonen-Sinisalo L, Sampson JR, Stormorken A, Wijnen J (2007) Guidelines for the clinical management of Lynch syndrome (hereditary non-polyposis cancer). J Med Genet 44(6):353–362
de la Chapelle A (2005) The incidence of Lynch syndrome. Fam Cancer 4(3):233–237
Maquat LE (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5(2):89–99
Naruse H, Ikawa N, Yamaguchi K, Nakamura Y, Arai M, Ishioka C, Sugano K, Tamura K, Tomita N, Matsubara N, Yoshida T, Moriya Y, Furukawa Y (2009) Determination of splice-site mutations in Lynch syndrome (hereditary non-polyposis colorectal cancer) patients using functional splicing assay. Fam Cancer 8(4):509–517
Reese MG, Eeckman FH, Kulp D, Haussler D (1997) Improved splice site detection in Genie. J Comput Biol 4(3):311–323
Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37(9):e67
Ng PC, Henikoff S (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31(13):3812–3814
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248–249
Vasen HF, Mecklin JP, Khan PM, Lynch HT (1991) The international collaborative group on hereditary non-polyposis colorectal cancer (ICG-HNPCC). Dis Colon Rectum 34(5):424–425
Genuardi M, Viel A, Bonora D, Capozzi E, Bellacosa A, Leonardi F, Valle R, Ventura A, Pedroni M, Boiocchi M, Neri G (1998) Characterization of MLH1 and MSH2 alternative splicing and its relevance to molecular testing of colorectal cancer susceptibility. Hum Genet 102(1):15–20
Sahashi K, Masuda A, Matsuura T, Shinmi J, Zhang Z, Takeshima Y, Matsuo M, Sobue G, Ohno K (2007) In vitro and in silicon analysis reveals an efficient algorithm to predict the splicing consequences of mutations at the 5′ splice sites. Nucleic Acids Res 35(18):5995–6003
Krawczak M, Thomas NS, Hundrieser B, Mort M, Wittig M, Hampe J, Cooper DN (2007) Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum Mutat 28(2):150–158
Lastella P, Resta N, Miccolis I, Quagliarella A, Guanti G, Stella A (2004) Site directed mutagenesis of hMLH1 exonic splicing enhancers does not correlate with splicing disruption. J Med Genet 41(6):e72
Auclair J, Busine MP, Navarro C, Ruano E, Montmain G, Desseigne F, Saurin JC, Lasset C, Bonadona V, Giraud S, Puisieux A, Wang Q (2006) Systematic mRNA analysis for the effect of MLH1 and MSH2 missense and silent mutations on aberrant splicing. Hum Mutat 27(2):145–154
Pagenstecher C, Wehner M, Friedl W, Rahner N, Aretz S, Friedrichs N, Sengteller M, Henn W, Buettner R, Propping P, Mangold E (2006) Aberrant splicing in MLH1 and MSH2 due to exonic and intronic variants. Hum Genet 119(1–2):9–22
Tournier I, Vezain M, Martins A, Charbonnier F, Baert-Desurmont S, Olschwang S, Wang Q, Buisine MP, Soret J, Tazi J, Frebourg T, Tosi M (2008) A large fraction of unclassified variants of the mismatch repair genes MLH1 and MSH2 is associated with splicing defects. Hum Mutat 29(12):1412–1424
Wagner A, Barrows A, Wijnen JT, van der Klift H, Franken PF, Verkuijlen P, Nakagawa H, Geugien M, Jaghmohan-Changur S, Breukel C, Meijers-Heijboer H, Morreau H, van Puijenbroek M, Burn J, Coronel S, Kinarski Y, Okimoto R, Watson P, Lynch JF, de la Chapelle A, Lynch HT, Fodde R (2003) Molecular analysis of hereditary nonpolyposis colorectal cancer in the United States: high mutation detection rate among clinically selected families and characterization of an American founder genomic deletion of the MSH2 gene. Am J Hum Genet 72(5):1088–1100
Zhao Y, Miyashita K, Ando T, Kakeji Y, Yamanaka T, Taguchi K, Ushijima T, Oda S, Maehara Y (2008) Exclusive KRAS mutation in microsatellite-unstable human colorectal carcinomas with sequence alterations in the DNA mismatch repair gene, MLH1. Gene 423(2):188–193
Giraldo A, Gomez A, Salguero G, Garcia H, Aristizabal F, Gutierrez O, Angel LA, Padron J, Martinez C, Martinez H, Malaver O, Florez L, Barvo R (2005) MLH1 and MSH2 mutations in Colombian families with hereditary nonpolyposis colorectal cancer (Lynch syndrome)–description of four novel mutations. Fam Cancer 4(4):285–290
Ewald J, Rodrigue CM, Mourra N, Lefevre JH, Flejou JF, Tiret E, Gespach C, Parc YR (2007) Immunohistochemical staining for mismatch repair proteins, and its relevance in the diagnosis of hereditary non-polyposis colorectal cancer. Br J Surg 94(8):1020–1027
Shin YK, Heo SC, Shin JH, Hong SH, Ku JL, Yoo BC, Kim IJ, Park JG (2004) Germline mutations in MLH1, MSH2 and MSH6 in Korean hereditary non-polyposis colorectal cancer families. Hum Mutat 24(4):351
Kohonen-Corish M, Ross VL, Doe WF, Kool DA, Edkins E, Faragher I, Wijnen J, Khan PM, Macrae F, St John DJ (1996) RNA-based mutation screening in hereditary nonpolyposis colorectal cancer. Am J Hum Genet 59(4):818–824
Bianchi F, Raponi M, Piva F, Viel A, Bearzi I, Galizia E, Bracci R, Belvederesi L, Loretelli C, Brugiati C, Corradini F, Baralle D, Cellerino R (2011) An intronic mutation in MLH1 associated with familial colon and breast cancer. Fam Cancer 10(1):27–35
Yan SY, Zhou XY, Du X, Zhang TM, Lu YM, Cai SJ, Xu XL, Yu BH, Zhou HH, Shi DR (2007) Three novel missense germline mutations in different exons of MSH6 gene in Chinese hereditary non-polyposis colorectal cancer families. World J Gastroenterol 13(37):5021–5024
Raevaara TE, Korhonen MK, Lohi H, Hampel H, Lynch E, Lonnqvist KE, Holinski-Feder E, Sutter C, McKinnon W, Duraisamy S, Gerdes AM, Peltomaki P, Kohonen-Ccorish M, Mangold E, Macrae F, Greenblatt M, de la Chapelle A, Nystrom M (2005) Functional significance and clinical phenotype of nontruncating mismatch repair variants of MLH1. Gastroenterology 129(2):537–549
Takahashi M, Shimodaira H, Andreutti-Zaugg C, Iggo R, Kolodner RD, Ishioka C (2007) Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res 67(10):4595–4604
Kansikas M, Kariola R, Nystrom M (2011) Verification of the three-step model in assessing the pathogenicity of mismatch repair gene variants. Hum Mutat 32(1):107–115
Acknowledgments
This study was supported in part by Ministry of Education, Science, Sports and Culture.
Conflict of Interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
This study is contacted for HNPCC registry and genetic testing project of the Japanese Society for Cancer of the Colon and Rectum (JSCCR).
Rights and permissions
About this article
Cite this article
Takahashi, M., Furukawa, Y., Shimodaira, H. et al. Aberrant splicing caused by a MLH1 splice donor site mutation found in a young Japanese patient with Lynch syndrome. Familial Cancer 11, 559–564 (2012). https://doi.org/10.1007/s10689-012-9547-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-012-9547-1