Abstract
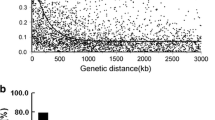
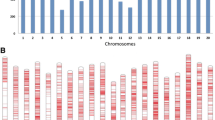
Damage generated by insects is one of main restricting factors for soybean production. Stink bugs are a great threat within pests because, by feeding mainly on pods, they cause direct and irreversible damage to developing seeds. Thus, plant resistance is an important management strategy to reduce insect population impact on yield losses. Association mapping can be used as a powerful tool for dissecting resistance mechanisms in soybean, more specifically to recover functional loci involved in plant defense against herbivorous insects; and can also provide valuable markers for the development of soybean cultivars with resistance. The purpose of this study was to identify molecular markers associated with resistance genes to stink bugs in a collection of soybean germplasm, using the association mapping strategy. According to the decline value in the linkage disequilibrium, an accurate power of mapping resolution was predicted in this population. Four associated markers located in chromosomes 6 and 15 were identified. Out of the 112 candidate genes close to them, 31 would encode proteins related to defense pathways triggered by the attack of herbivorous insects. The proteins encoded by these candidate genes could be associated to the jasmonic acid pathway. The main contribution of this study was the identification of molecular markers associated with resistance genes to stink bugs. These markers will be a useful tool for marker-assisted selection applied to soybean genetic breeding.





Similar content being viewed by others
References
Ainsworth EA, Yendrek CR, Skoneczka JA, Long SP (2012) Accelerating yield potential in soybean: potential targets for biotechnological improvement. Plant Cell Environ 35:38–52. https://doi.org/10.1111/j.1365-3040.2011.02378.x
Bansal R, Jun T, Mian M, Michel AP (2013) Developing host-plant resistance for hemipteran soybean pests: lessons from soybean aphid and stink bugs. Soybean Pest Resist 19:46. https://doi.org/10.5772/54597
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Bradbury P, Parker T, Hamblin MT, Jannink JL (2011) Assessment of power and false discovery rate in genome-wide association studies using the BarleyCAP germplasm. Crop Sci 51(1):52–59
Chang HX, Hartman GL (2017) Characterization of insect resistance loci in the USDA soybean germplasm collection using genome-wide association studies. Front Plant Sci 8:670. https://doi.org/10.3389/fpls.2017.00670
Chang HX, Lipka AE, Domier LL, Hartman GL (2016) Characterization of disease resistance loci in the USDA soybean germplasm collection using genome-wide association studies. Phytopathology 106(10):1139–1151. https://doi.org/10.1094/PHYTO-01-16-0042-FI
Di Rienzo JA, Casanoves F, Balzarini MG, Gonzalez L, Tablada M, Robledo CW (2016) InfoStat versión 2016. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. http://www.infostat.com.ar
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361. https://doi.org/10.1007/s12686-011-9548-7
Ehrenreich IM, Stafford PA, Purugganan MD (2007) The genetic architecture of shoot branching in Arabidopsis thaliana: a comparative assessment of candidate gene associations vs. quantitative trait locus mapping. Genetics 176(2):1223–1236
Ersoz ES, Yu J, Buckler ES (2008) Applications of linkage disequilibrium and association mapping in crop plants. In: Varshney R, Tuberosa R (eds) Genomic assisted crop improvement, vol I. Genomics approaches and platforms. Springer Verlag, Germany, pp 97–120
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinf 1:47–50. https://doi.org/10.1177/117693430500100003
Fehr W (1991) Principles of cultivar development: theory and technique. Macmillian Publishing Company, New York
Ghione C (2008) Evaluación de la diversidad genética entre genotipos de soja mediante la utilización de marcadores moleculares. Thesis, Universidad Nacional de Buenos Aires
Giorda LM, Baigorri H (1997) El Cultivo de la Soja en Argentina. INTA, Buenos Aires
Hanson AA, Lorenz AJ, Hesler LS, Bhusal SJ, Bansal R, Michel AP, Jiang G-L, Koch RL (2018) Genome-wide association mapping of host-plant resistance to soybean aphid. Plant Gen 11:3. https://doi.org/10.3835/plantgenome2018.02.0011
Hou J, Wang C, Hong X, Zhao J, Xue C, Guo N et al (2011) Association analysis of vegetable soybean quality traits with SSR markers. Plant Breed 130(4):444–449. https://doi.org/10.1111/j.1439-0523.2011.01852.x
Hymowitz T (2004) Speciation and cytogenetics. In: Boerma HR, Specht JE (eds) Soybeans: Improvement, Production and Uses. American Society of Agronomy, Crop Science Society of America, Soil Science Society of America, Madison, pp 97–136
Hymowitz T, Newell CA (1981) Taxonomy of the genus Glycine, domestication and uses of soybeans. Econ Bot 35:272–288. https://doi.org/10.1007/BF02859119
Hyten DL, Choi IY, Song Q, Shoemaker RC, Nelson RL, Costa JM et al (2007) Highly variable patterns of linkage disequilibrium in multiple soybean populations. Genetics 175(4):1937–1944. https://doi.org/10.1534/genetics.106.069740
Kim H, Xing G, He J, Zhao T, Yang S, Li Y, Palmer RG, Gai J (2015) An environmental differential association analysis of antibiosis to common cutworm in a Chinese soybean germplasm population and optimization of the cross design. Mol Breed 35(2):76. https://doi.org/10.1007/s11032-015-0267-8
Kim H, Xing G, Wang Y, Zhao T, Yu D, Yang S, Li Y, Chen S, Palmer RG, Gai J (2014) Constitution of resistance to common cutworm in terms of antibiosis and antixenosis in soybean RIL populations. Euphytica 196(1):137–154. https://doi.org/10.1007/s10681-013-1021-0
Kloth KJ, Thoen MP, Bouwmeester HJ, Jongsma MA, Dicke M (2012) Association mapping of plant resistance to insects. Trends Plant Sci 17(5):311–319. https://doi.org/10.1016/j.tplants.2012.01.002
Lee JS, Yoo MH, Jung JK, Bilyeu KD, Lee JD, Kang S (2015) Detection of novel QTLs for foxglove aphid resistance in soybean. Theor Appl Genet 128(8):1481–1488. https://doi.org/10.1007/s00122-015-2519-8
Li Y, Guan R, Liu Z, Ma Y, Wang L, Li L et al (2008) Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor Appl Genet 117(6):857–871. https://doi.org/10.1007/s00122-008-0825-0
Liu H, Che Z, Zeng X, Zhang G, Wang H, Yu D (2016) Identification of single nucleotide polymorphisms in soybean associated with resistance to common cutworm (Spodoptera litura Fabricius). Euphytica 209(1):49–62. https://doi.org/10.1007/s10681-016-1631-4
Locatelli A, Cuesta-Marcos A, Gutiérrez L, Hayes PM, Smith KP, Castro AJ (2013) Genome-wide association mapping of agronomic traits in relevant barley germplasm in Uruguay. Mol Breed 31(3):631–654
Mao T, Li J, Wen Z, Wu T, Wu C, Sun S et al (2017) Association mapping of loci controlling genetic and environmental interaction of soybean flowering time under various photo-thermal conditions. BMC Genom 18(1):415. https://doi.org/10.1186/s12864-017-3778-3
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8(19):4321–4326. https://doi.org/10.1093/nar/8.19.4321
Panizzi AR, Slansky F Jr (1985) Review of phytophagous pentatomids (Hemiptera: Pentatomidae) associated with soybean in the Americas. Florida Entomol 1:184–214. https://doi.org/10.2307/3494344
Priolli RHG, Campos JB, Stabellini NS, Pinheiro JB, Vello NA (2015) Association mapping of oil content and fatty acid components in soybean. Euphytica 203(1):83–96. https://doi.org/10.1007/s10681-014-1264-4
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Salvi S, Sponza G, Morgante M, Tomes D, Niu X, Fengler KA, Salvi S, Sponza G, Morgante M, Tomes D, Niu X, Fengler KA, Meeley R, Ananiev EV, Svitashev S, Bruggemann EB, Li B (2007) Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci 104(27):11376–11381
Santos MDF (2012) Mapeamento de QTL e expressão gênica associados à resistência da soja ao complexo de percevejos. Doctoral dissertation, Universidade de São Paulo.
Van Os H, Andrzejewski S, Bakker E, Barrena I, Bryan GJ, Caromel B, GhareebIsidore B, DeJong E, Van Koert W, Lefebvre PV (2006) Construction of a 10,000-marker ultradense genetic recombination map of potato: providing a framework for accelerated gene isolation and a genomewide physical map. Genetics 173(2):1075–1087
Wang LX, Guan RX, Li YH, Lin FY, Luan WJ, Li W et al (2008) Genetic diversity of Chinese spring soybean germplasm revealed by SSR markers. Plant Breed 127(1):56–61. https://doi.org/10.1111/j.1439-0523.2007.01429.x
Wang H, Yan H, Du H, Chao M, Gao Z, Yu D (2015) Mapping quantitative trait loci associated with soybean resistance to common cutworm and soybean compensatory growth after defoliation using SNP marker-based genome-wide association analysis. Mol Breed 35(8):168. https://doi.org/10.1007/s11032-015-0360-z
Wen Z, Tan R, Yuan J, Bales C, Du W, Zhang S et al (2014) Genome-wide association mapping of quantitative resistance to sudden death syndrome in soybean. BMC Genom 15(1):809. https://doi.org/10.1186/1471-2164-15-809
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ghione, C.E., Lombardo, L.A., Vicentin, I.G. et al. Association mapping to identify molecular markers associated with resistance genes to stink bugs in soybean. Euphytica 217, 46 (2021). https://doi.org/10.1007/s10681-021-02768-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-021-02768-1