Abstract
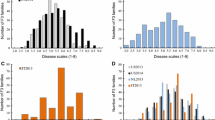
Hop with powdery mildew [HPM: caused by Podosphaera macularis (Wallr.) U. Braun & S. Takam.] results in significant losses in hop production by reducing yield and quality. One means of increasing yield and quality is the production of resistant hop lines. Breeding for resistance can be significantly improved and accelerated by use of marker-assisted selection. The objective of this preliminary study was to identify QTLs and markers for genetic resistance to HPM. A bi-parental mapping population between the resistant line “Newport” and susceptible line ‘21110M’. Phenotypic data was scored under controlled greenhouse conditions. Significant differences among offspring were observed and disease resistance did not follow a distinct binomial distribution, suggesting quantitative genetic control. Genotyping-by-sequencing resulted in approximately 375 K SNP markers, which were filtered down to 2263 markers mapped to 10 linkage groups. Interval Mapping identified four QTLs with one on linkage group 1 and three located on linkage group 6. Composite interval mapping identified three QTLs, all located on linkage group 6. Mixed linear models identified 15 markers associated with expression of resistance to HPM. Three of these 15 SNPs were also identified in QTL-CIM analysis. Evaluation of the scaffolds containing the significant SNP markers identified seven putative genes—several of which appear involved in disease resistance in other plant species. The SNP markers identified in this study still require validation in unrelated populations prior to implementation in breeding programs.





Similar content being viewed by others
References
Allard R (1999) Principles of plant breeding. Wiley: Hoboken. ISBN: 978-0-471-02309-8
Austin MB, Noel JP (2003) The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep 20:79–110
Ben-Ari G, Lavi U (2012) 11-marker-assisted selection in plant breeding A2—Altman, Arie. In: Hasegawa PM (ed) Plant biotechnology and agriculture. Academic Press, San Diego, pp 163–184
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57 (1): 289–300. MR 1325392
Bernardo R (2008) Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci 48:1649–1664
Bernardo R (2010) Breeding for quantitative traits in plants. Stemma Press, Woodbury
Bonferroni CE (1936) “Teoria statistica delle classi e calcolo delle probabilità.”. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze 8: 3–62,
Borner GH, Lilley KS, Stevens TJ, Dupree P (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol 132:568–577
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Castro CB, Whittock LD, Whittock SP, Leggett G, Koutoulis A (2008) DNA sequence and expression variation of Hop (Humulus lupulus) valerophenone synthase (VPS), a key gene in bitter acid biosynthesis. Ann Bot 102:265–273
Čerenak A, Šatović Z, Jakše J, Luthar Z, Carović-Stanko K, Javornik B (2009) Identification of QTLs for alpha acid content and yield in hop (Humulus lupulus L.). Euphytica 170(1–2):141–154. doi:10.1007/s10681-009-9920-9
Dao TTH, Linthorst HJM, Verpoorte R (2011) Chalcone synthase and its functions in plant resistance. Phytochem Rev 10:397–412
Djian-Caporalino C, Palloix A, Fazari A, Marteu N, Barbary A, Abad P, Sage-Palloix A-M, Mateille T, Risso S, Lanza R et al (2014) Pyramiding, alternating or mixing: comparative performances of deployment strategies of nematode resistance genes to promote plant resistance efficiency and durability. BMC Plant Biol 14:1–13
Dudley JW (2007) From means to QTL: the Illinois long-term selection experiment as a case study in quantitative genetics. Crop Sci 47(S3):S20–S31
Flor HH (1971) Current status of the gene-for-gene concept. Ann Rev Phytopathol 9:275–296
Gent DH, Nelson ME, George AE, Grove GG, Mahaffee WF, Ocamb CM, Barbour JD, Peetz A, Turechek WW (2008) A decade of hop powdery mildew in the pacific northwest. Plant Health Prog. doi:10.1094/PHP-2008-0314-01-RV
Godwin JR, Mansfield JW, Darby P (1987) Microscopical studies of resistance to powdery mildew disease in the hop cultivar Wye Target. Plant Pathol 36:21–32
Hallauer AR (2007) History, contribution, and future of quantitative genetics in plant breeding: lessons from maize. Crop Sci 47(S3):S4–S19
Henning JA, Townsend MS, Mahaffee WF, Kenny S, Haunold A (2004) Registration of ‘Newport’ hop. Crop Sci 44:1018–1019
Henning J, Townsend MS, Gent D, Bassil N, Matthews P, Buck E, Beatson R (2011) QTL mapping of powdery mildew susceptibility in hop (Humulus lupulus L.). Euphytica 180:411–420
Henning JA, Gent DH, Twomey MC, Townsend MS, Pitra NJ, Matthews PD (2015) Precision QTL mapping of downy mildew resistance in hop (Humulus lupulus L.). Euphytica 202:487–498
Henning JA, Gent DH, Twomey MC, Townsend MS, Pitra NJ, Matthews PD (2016). Genotyping-by-sequencing of a bi-parental mapping population segregating for downy mildew resistance in hop (Humulus lupulus L.). Euphytica 208:545–559. doi:10.1007/s10681-015-1600-3
Hill, ST, JA. Henning, R Sudarsanam, and D Hendrix. (2016). HopBase: A unified resource for Humulus Genomics. J. Database (Submitted)
Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 121:185–199
Li F, Zhang H, Wang S, Xiao W, Ding C, Liu W, Guo H (2016) Identification of topping responsive proteins in tobacco roots. Front Plant Sci 7:582
Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH et al (2013) Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genet 9(1):e1003215. doi:10.1371/journal.pgen.1003215
Mahaffee WF, Thomas CS, Turechek WW, Ocamb CM, Nelson ME, Fox A, Gubler WD (2003) Responding to an introduced pathogen: Podosphaera macularis (hop powdery mildew) in the Pacific Northwest. Plant Health Prog. doi:10.1094/PHP-2003-1113-07-RV
Mahaffee WF, Engelhard B, Gent DH, Groves GG (2009) Powdery mildew. In: Mahaffee WF, Pethybridge SJ, Gent DH (eds) Compendium of hop diseases and pests. American Phytopathological Society Press, St. Paul
McAdam EL, Freeman JS, Whittock SP, Buck EJ, Jakse J, Cerenak A, Javornik B, Kilian A, Wang C-H, Andersen D et al (2013) Quantitative trait loci in hop (Humulus lupulus L.) reveal complex genetic architecture underlying variation in sex, yield and cone chemistry. BMC Genom 14:1–27
Nambiar M, Smith GR (2016) Repression of harmful meiotic recombination in centromeric regions. Semin Cell Dev Biol 54:188–197
Natsume S, Takagi H, Shiraishi A, Murata J, Toyonaga H, Patzak J, Takagi M, Yaegashi H, Uemura A, Mitsuoka C et al (2015) The draft genome of hop (Humulus lupulus), an essence for brewing. Plant Cell Physiol 56:428–441
Neve RA (1991) Hops. Chapman & Hall, London
Ocamb CM, Gent DH (2016) Hop (Humulus lupulus)-Powdery mildew. In: Pscheidt JW, Ocamb CM (eds.) Pacific northwest plant disease management handbook . Corvallis, OR: Oregon State University. http://pnwhandbooks.org/plantdisease/hop-humulus-lupulus-abiotic-wilt. Accessed 31 March 2016
Poland J, Rutkoski J (2016) Advances and challenges in genomic selection for disease resistance. Annu Rev Phytopathol 54:79–98
Reilly JF, Martinez SD, Mickey G, Maher PA (2002) A novel role for farnesyl pyrophosphate synthase in fibroblast growth factor-mediated signal transduction. Biochem J 366:501–510. doi:10.1042/bj20020560
Sallam AH, Smith KP (2016) Genomic selection performs similarly to phenotypic selection in barley. Crop Sci 56:2871–2881. doi:10.2135/cropsci2015.09.0557
St. Clair DA (2010) Quantitative disease resistance and quantitative resistance loci in breeding. Annu Rev Phytopathol 48:247–268
Van Ooijen J (2011) Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet Res 93(5):343–349
Wang S, Basten CJ, Zeng ZB (2012) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm. Accessed 9 Oct 2016
Wolfenbarger SN, Eck EB, Gent DH (2014) Characterization of resistance to powdery mildew in the hop cultivars “Newport” and “Comet”. Plant Health Prog. doi:10.1094/PHP-BR-13-0129
Zimmermann CE, Likens ST, Haunold A, Homer CE, Roberts DD (1975) Registration of “Comet” hop (Registration No. 3). Crop Sci 15:98
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Henning, J.A., Gent, D.H., Townsend, M.S. et al. QTL analysis of resistance to powdery mildew in hop (Humulus lupulus L.). Euphytica 213, 98 (2017). https://doi.org/10.1007/s10681-017-1849-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-017-1849-9