Abstract
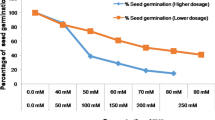
To expand the salt-tolerant gene resources of peanut, we conducted in vitro mutagenesis with pingyangmycin (PYM) as the mutagen and directed screening with medium containing NaCl. After embryonic leaflets from mature peanut seeds (variety Huayu 22) were cultured on somatic embryogenesis-induction medium containing 4 mg/L PYM for 4 weeks, the surviving somatic embryos were sequentially transferred to a germination medium containing 15 and then 20 g/L NaCl. The 30 NaCl-tolerant plantlets obtained were grafted and transplanted in the field in 2011, and the mature seeds of 26 regenerated plants were harvested. In 2012, all seeds from each plant were sown in the field. The offspring (M2 generation) of 23 of 26 NaCl-tolerant, regenerated plants differed from their mutagenic parent in vigor, growth habit, flowering habit, pod shape, and seed coat color, and they also exhibited trait segregation from the same NaCl-tolerant, regenerated plant. In a germination test with a 0.7 % NaCl solution and the M3-generation seeds from 18 of the NaCl-tolerant, regenerated plants, the germination rate was substantially higher for the seeds from 6 plants than for seeds from the mutagenic parent (Huayu 22). To determine whether the changes in plant traits might be associated with gene mutations, DNA polymorphisms between the mutagenic parent and 19 M3 generation individuals from different NaCl-tolerant, regenerated plants were analyzed with 39 pairs of SSR primers, and all mutants differed from the mutagenic parent in >2 loci. The results indicate that the use of PYM-based mutagenesis in combination with directed in vitro screening with NaCl is effective for creating and identifying salt-tolerant mutants of peanut.







Similar content being viewed by others
References
Ahloowalia BS, Maluszynski M, Nichterlein K, van Zanten L, Weck E (1998) Induced mutations and in vitro culture techniques for the improvement of horticultural plants. In: Chopra VL, Singh RB, Varma A (eds) Crop productivity and sustainability: shaping the future. Oxford and IBH, New Delhi, pp 405–412
Dita MA, Rispail N, Prats E, Rubiales D, Singh KB (2006) Biotechnology approaches to overcome biotic and abiotic stress constraints in legumes. Euphytica 147:1–24
Fuller MP, Metwali EMR, Eed MH, Jellings AJ (2006) Evaluation of abiotic stress resistance in mutated populations of cauliflower (Brassica oleracea var. botrytis). Plant Cell Tiss Org Cult 86:239–248
Gowda MVC, Nadaf HL, Sheshagiri R (1996) The role of mutations in intraspecific differentiation of groundnut (Arachis hypogaea L.). Euphytica 90:105–113
Hu XH, Sun LQ, Miao HR, Shi YQ, Chen J (2011) Effects of different NaCl concentrations on indicators for evaluating salt tolerance of peanut varieties. J Shandong Agric Sci 11:35–37
Jain SM (2001) Tissue culture-derived variation in crop improvement. Euphytica 118:153–166
Lee JH, Lee SY (2002) Selection of stable mutants from cultured rice anthers treated with ethyl methane sulfonic acid. Plant Cell Tiss Organ Cult 71(2):165–171
Li AX, Liu QC, Wang YP, Zhai H, Wang SF, Liu BL (2002) In vitro selection of drought and salt tolerant mutants in sweet potato. J Agric Biotechnol 10(1):15–19
Liu JP, Zhen CM (2004) Induced mutation in connection with in vitro culture for crop breeding. Acta Agric Shanghai 20(1):19–22
Liu YH, Shen Y, Wang ZF, Yan W (2012) Identification of salt tolerance in peanut varieties/lines at the germination stage. Chin J Oil Crop Sci 34(2):168–173
Luan YS, Zhang J, Gao XR (2007) Mutation induced by ethylmethanesulfonate (EMS), in vitro screening for salt tolerance and plant regeneration of sweet potato (Ipomoae batatas L.). Plant Cell Tiss Org Cult 88:77–81
Milla SR, Isleib TG, Stalker HT (2005) Taxonomic relationships among Arachis sect. Arachis species as revealed by AFLP markers. Genome 48:1–11
Muthusamy A, Vasanth K, Sivasankari D, Chandrasekar BR, Jayabalan N (2007) Effects of mutagens on somatic embryogenesis and plant regeneration in groundnut. Biol Plant 51(3):430–435
Patade VY, Suprasanna P (2009) An in vitro radiation induced mutagenesis-selection system for salinity tolerance in sugarcane. Sugar Tech 11(3):246–251
Predieri S (2001) Mutation induction and tissue culture in improving fruits. Plant Cell Tiss Org Cult 64:185–210
Rai MK, Kalia RK, Singh R, Gangola MP, Dhawan AK (2011) Developing stress tolerant plants through in vitro selection-an overview of the recent progress. Environ Exp Bot 71:89–98
Shirasawa K, Koilkonda P, Aoki K, Hirakawa H, Tabata S, Watanabe M, Hasegawa M, Kiyoshima H, Suzuki S, Kuwata C, Naito Y, Kuboyama T, Nakaya A, Sasamoto S, Watanabe A, Kato M, Kawashima K, Kishida Y, Kohara M, Kurabayashi A, Takahashi C, Tsuruoka H, Wada T, Isobe S (2012) In silico polymorphism analysis for the development of simple sequence repeat and transposon markers and construction of linkage map in cultivated peanut. BMC Plant Biol 12:80
Sui JM, Li R, Fan QC, Song L, Zheng CH, Wang JS, Qiao LX, Yu SL (2013) Isolation and characterization of a stress responsive small GTP-binding protein AhRabG3b in peanut (Arachis hypogaea L.). Euphytica 189:161–172
Sun GZ, Tang FL, Wang GJ, Li ZJ, Zhang YM, Yan WY, Sun DQ, Sun Y (1998) Radiation induced and biological technology in combination with create new wheat qualitative research. J Tritical Crops 18(6):1–4
Venkatachalam P, Jayabalam N (1997) Selection and regeneration of groundnut plants resistant to the pathotoxic culture filtrate of Cercosporidium personation through tissue culture technology. Appl Biochem Biotechnol 61(3):351–364
Wang H, Chen FL, Zhao HB, Fang WM (2007) Mutagenic effect of Pingyangmycin on Dendranthema grandiflorum ‘Italy Red’ with small inflorescence. J Nanjing Agric Univ 30(1):39–45
Wu GT, Xia YW, Shu QY, Wang XY, Zhou Y (1997) Mutagenic effects of pingyangmycin on rice (Oryza sativa L.). J Zhejiang Agric Univ 23(1):1–6
Wu LR, Chen J, Xu TT, Miao HR, Hu WG, Yu SL (2005a) Identification of salt tolerance in peanut growth duration. J Peanut Sci 34(1):20–24
Wu WG, Liu JR, Yang XJ (2005b) Applications of the mutation in connection with in vitro culture for plant breeding. J China Agric 21:197–201
Wynne JC, Coffelt TA (1982) Genetics of Arachis hypogaea L. In: Pattee HE, Young CT (eds) Peanut science and technology. APRES, Yoakum/Ty, pp 50–94
Xu SB, Tao YF, Yang ZQ, Chu JY (2002) A simple and rapid method used for silver staining and gel preservation. Hereditas 24(3):335–336
Yang Q, Zhang F, Wang D, Zhang JL, Yang HY, Liu YH (2011) Selection of salt-tolerant variants from potato in vitro micro-cuttings induced by ems. J Tritieeae Crops 4:679–683
Zhang XT (2011) Study on character variation of ‘Lankaoaiza08’ induced by pingyangmycin and assessment of genetic diversity of the descendant using SSR markers. J Chin Agric Sci Bull 27(11):36–41
Zhang QY, Ye DS, Jin MY (2002) Studies on variations of wheat agronomic characters induced by Pingyangmycin. J Tritieeae Crops 22(4):66–69
Zhao MX, Sui JM, Wang XJ, Qiao LX, Guo BT, Wang JS (2011) Effect of pingyangmycin on somatic embryogenesis in embryonic leaflet culture of peanut. J Nuclear Agri Sci 25(2):0242–0246
Zhou LM, Wang F, Wang J (2009) Selection of salt-tolerant mutants from ethyl methane sulfonate mutagenized kiwifruit embryonic calli. J Acta Agriculturae Boreali-occidentalis Sinica 18(5):330–335
Acknowledgments
This study was financially supported by the National Natural Science Foundation of the P. R. China (31101178), Generation of Mutant Pool of Peanut, Gene mining, Germplasm Innovation and Utilization Project of Shandong, and the Natural Science Foundation of Shandong (ZR2012CM014 and ZR2011CQ026).
Author information
Authors and Affiliations
Corresponding author
Additional information
Ming-Xia Zhao, Hai-Yan Sun and Rui-Rui Ji have contributed equally to this study.
Rights and permissions
About this article
Cite this article
Zhao, MX., Sun, HY., Ji, RR. et al. In vitro mutagenesis and directed screening for salt-tolerant mutants in peanut. Euphytica 193, 89–99 (2013). https://doi.org/10.1007/s10681-013-0956-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-013-0956-5