Abstract
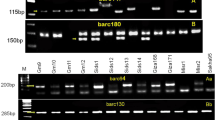
Late leaf spot (LLS) caused by Phaeoisariopsis personata is the major foliar disease that reduces the pod yield and severely affects the fodder and seed quality in groundnut. Molecular markers linked with LLS can improve the process of identification of resistant genotypes. In the present study, a LLS susceptible genotype (TMV 2) and the LLS resistant genotype (COG 0437) were crossed and their F2 population was used for marker analysis. The phenotypic mean data on F2:3 progenies were used as phenotype. Parents were surveyed with 77 SSR (Simple Sequence Repeat) primers to identify polymorphic markers. Among SSR markers, nine primers were found polymorphic between the parents TMV 2 and COG 0437. These markers were utilized for bulked segregant analysis (BSA). Among the polymorphic SSR markers, three primers viz., PM 375162, pPGPseq5D5220 and PM 384100 were able to distinguish the resistant and susceptible bulks and individuals for LLS. In single marker analysis, the markers PM 375, PM 384, pPGPseq5D5, PM 137, PM 3, PMc 588 and Ah 4-26 were linked with LLS severity score. The phenotypic variation explained by these markers ranged from 32 to 59 %. The markers identified through BSA were also confirmed with single marker analysis. While validating the three primers over a set of resistant and susceptible genotypes, the primer PM 384100 allele had association with resistance. Hence PM 384 could be utilized in the marker assisted breeding programme over a wide range of genetic background.

Similar content being viewed by others
References
Anderson WF, Wynne JC, Green CC (1986) Potential for incorporation of early and late leaf spot resistance in peanut. Plant Breed 97:163–170
Anonymous (2008) Agricultural Statistics at a Glance-2007-08. Directorate of Economics and Stastistics, Ministry of Agriculture, Government of India, New Delhi
Benbouza H, Jacquemin JM, Baudoin JP, Mergeai G (2006) Optimization of a reliable, fast, cheap and sensitive silver staining method to detect SSR markers in polyacrylamide gels. Biotechnol Agron Soc Environ 10(2):77–81
Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK (2005) An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142(1–2):169–196
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Dudley JW (1993) Molecular markers in plant improvement: manipulation of genes affecting quantitative traits. Crop Sci 33:660–668
Dwivedi SL, Gurtu S, Chandra S, Yuejin W, Nigam SN (2001) Assessment of genetic diversity among selected groundnut germplasm. I: RAPD analysis. Plant Breed 120:345–349
Dwivedi SL, Pande S, Rao JN, Nigam SN (2002) Components of resistance to late leaf spot and rust among interspecific derivatives and their significance in a foliar disease resistance breeding in groundnut (Arachis hypogaea L.). Euphytica 125:81–88
Food and Agriculture Organization of the United Nations (FAOSTAT) (2007) http://faostat.fao.org/site/567/default.aspx. Accessed 30 Mar 2009
Green CC, Wynne JC (1986) Diallel and generation means analyses for the components of resistance to Cercospora arachidicola in peanut. Theor Appl Genet 73:228–235
Green CC, Wynne JC (1987) Genetic variability and heritability for resistance to early leaf spot in four crosses of Virginia-type peanut. Crop Sci 27:18–21
Guang C, Xiong-Ming DU (2006) Genetic diversity source germplasm of upland cotton in China as determined by SSR marker analysis. Acta Genet Sin 33:733–745
Halward TM, Stalker HT, LaRue EA, Kochert G (1991) Genetic variation detectable with molecular markers among unadapted germplasm resources of cultivated peanut and related wild species. Genome 34:1013–1020
Hamid MA, Isleib TG, Wynne JC, Green CC (1981) Combining ability analysis of Cercospora leaf spot resistance and agronomic traits in Arachis hypogaea L. Oleagineux 36:605–612
He G, Prakash CS (2001) Evaluation of genetic relationships among botanical varieties of cultivated peanut (Arachis hypogaea L.) using markers. Genet Resour Crop Evol 48:347–352
Jogloy S, Wynne JC, Beute MK (1987) Inheritance of late leaf spot resistance and agronomic traits in peanut. Peanut Sci 14:86–90
Kornegay JL, Beute MK, Wynne JC (1980) Inheritance of resistance to Cercospora arachidicola and Cercosporidium personatum in six Virginia-type peanut lines. Peanut Sci 7:4–9
Krishna GK, Zhang JF, Burow M, Pittman RN, Lu Y, Puppala N, Elikostadinov SG (2004) Genetic diversity analysis in Valencia peanut (Arachis hypogaea L.) using microsatellite markers. Cell Mol Biol Lett 9(4):685–697
Leal-Bertioli SCM, Jose ACVF, Alves-Freitas DMT, Moretzsohn MC, Guimaraes PM, Nielen S, Vidigal BS, Pereira RW, Pike J, Favero AP, Parniske M, Varshney RK, Bertioli DJ (2009) Identification of candidate genome regions controlling disease resistance in Arachis. BMC Plant Biol 9:112–123
Li YC, Fahima T, Roder MS, Kirzhner VM, Beiles A, Korol AB, Nevo E (2003) Genetic effects on microsatellite diversity in wild emmer wheat (Triticum dicoccoides) at the Yehudiyya microsite, Israel. Heredity 90:150–156
Mace ES, Phong DT, Upadhaya HD, Chandra S, Crouch JH (2006) SSR analysis of cultivated groundnut (Arachis hypogaea L.) germplasm resistant to rust and late leaf spot diseases. Euphytica 152(3):317–330
Michelmore RW, Paranand I, Kessele RV (1991) Identification of markers linked to disease resistance genes by bulk segregant analysis: a rapid method to detect markers in specific genome using segregant population. Proc Natl Acad Sci USA 88:9828–9832
Mondal S, Ghosh S, Badigannavar AM (2005) RAPD polymorphism among groundnut genotypes differing in disease reaction to late leaf spot and rust. Intl Arachis Newslett 25:27–30
Mondal S, Badigannavar AM, Murthy GSS (2007) RAPD markers linked to rust resistance in cultivated groundnut (Arachis hypogaea L.). Euphytica 159:233–239
Morgante M, Olivieri A (1993) PCR-amplified microsatellite as markers in plant genetics. Plant J 3:175–182
Motagi (2001) Genetic analysis of resistance to late leaf spot and rust vis-à-vis productivity in groundnut (Arachis hypogaea L.). Dissertation, University of Agriculture Sciences, Dharwad, India
Nevill DJ (1982) Inheritance of resistance to Cercosporidium personatum in groundnuts: a genetic model and its implications for selection. Oleagineux 37:355–362
Pande S, Rao NJ (2001) Resistance of wild Arachis species to late leaf spot and rust in greenhouse trials. Plant Dis 85:851–885
Raina SN, Rani V, Kojima T, Ojihara Y, Singh KP, Devarumath RM (2001) RAPD and ISSR fingerprints as useful genetic markers for analysis of genetic diversity, varietal identification, and phylogenetic relationships in peanut (Arachis hypogaea) cultivars and wild species. Genome 44(5):763–772
Ribaut JM, Banziger M, Betran J, Jiang C, Edmeades GO, Dreher K, Hoisington D (2002) Use of molecular markers in plant breeding: drought tolerance improvement in tropical maize. In: Kang MS (ed) Quantitative genetics genomics and plant breeding. CABI Publishing, Wallingford, pp 85–99
Sax K (1923) The association of size differences with seed coat pattern and pigmentation in Phaseolus vulgaris. Genetics 8:552–560
Selvaraj MG, Narayana M, Schubert AM, Ayers JL, Baring MR, Burow MD (2009) Identification of QTLs for pod and kernel traits in cultivated peanut by bulked segregant analysis. Electron J Biotechnol 12(2):13
Sharief Y, Rawlings JO, Gregory WC (1978) Estimates of leaf spot resistance in three interspecific hybrids of Arachis. Euphytica 27:741–751
Singh AK, Smartt J, Simpson CE, Raina SN (1998) Genetic variation vis-à-vis molecular polymorphism in groundnut, Arachis hypogaea L. Genet Resour Crop Evol 45:119–126
Subrahmanyam P, Reddy LJ, Gibbons RW, McDonald D (1985) Peanut rust: a major threat to peanut production in the semi arid tropics. Plant Dis 69:813–819
Subrahmanyam P, McDonald D, Waliyar F, Reddy LJ, Nigam SN, Gibbons RW, Rao VR, Singh AK, Pande S, Reddy PM, Subba Rao PV (1995) Screening methods and sources of resistance to rust and late leaf spot of groundnut. Information Bulletin No. 47, ICRISAT, Patancheru, p 24
Subramanian V, Gurtu S, Rao RCN, Nigam SN (2000) Identification of DNA polymorphism in cultivated groundnut using random amplified polymorphic DNA(RAPD) assay. Genome 43:656–660
Tang R, Gao G, He L, Han Z, Shan S, Zhong R, Zhou C, Jiang J, Li Y, Zhuang W (2007) Genetic diversity in cultivated groundnut based on SSR markers. J Genet Genomics 34(5):449–459
Tiwari SP, Ghewande MP, Misra DP (1984) Inheritance of resistance to rust and late leaf spot in groundnut (Arachis hypogaea). J Cytol Genet 19:97–101
Tommasini L, Bately J, Arnold GM, Cooke RJ, Donini P, Lee D, Law JR, Lowe C, Moule C, Trick M, Edwards KJ (2003) The development of multiplex simple sequence repeat (SSR) markers to complement distinctness, uniformity and stability testing of rape (Brassica napus L.) varieties. Theor Appl Genet 106:1091–1101
Varma TSN, Dwivedi SL, Pande S, Gowda MVC (2005) SSR markers associated with resistance to rust (Puccinia arachidis Speg.) in groundnut (Arachis hypogaea L.). SABRAO J 37(2):107–119
Varshney RK, Hoisington DA, Upadhyaya HD, Gaur PM, Nigam SN, Saxena K, Vadez V, Sethy NK, Bhatia S, Aruna R, Gowda MVC, Singh NK (2007) Molecular genetics and breeding of grain legume crops for the semi-arid tropics. In: Varshney RK, Tuberosa R (eds) Genomic assisted crop improvement genomics applications in crops. Springer, Dordrecht, pp 207–2420
Waliyar F (1991) Evaluation of yield losses due to groundnut leaf diseases in West Africa. In: Nduguru BJ, Waliyar F, Ntare BR (eds) Summary proceedings of the second ICRISAT regional groundnut meeting for West Africa. ICRISAT Sahelian Centre, Niamey, Niger, pp 32–33
Wang GL, Paterson AH (1994) Assessment of DNA pooling strategies for mapping of QTLs. Theor Appl Genet 88:355–361
Wang CT, Yang XD, Chen DX, Yu SL, Liu GZ, Tang YY, Xu JZ (2007) Isolation of simple sequence repeats from groundnut. Electron J Biotechnol 10(3):473–480
Wenzl P, Raman H, Wang J, Zhou M, Huttner E, Kilian A (2007) A DArT platform for quantitative bulked segregant analysis. BMC Genomics 8:196
Young ND, Weeden NF, Kochert G (1996) Genome mapping in legumes (Fam. Fabaceae). In: Paterson AH (ed) Genome mapping in plants. Landes Company, Austin, pp 211–227
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shoba, D., Manivannan, N., Vindhiyavarman, P. et al. SSR markers associated for late leaf spot disease resistance by bulked segregant analysis in groundnut (Arachis hypogaea L.). Euphytica 188, 265–272 (2012). https://doi.org/10.1007/s10681-012-0718-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-012-0718-9