Abstract
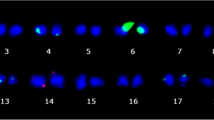
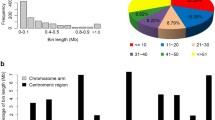
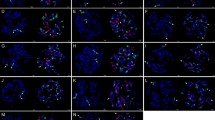
Molecular markers have been successfully used in rice breeding however available markers based on Oryza sativa sequences are not efficient to monitor alien introgression from distant genomes of Oryza. We developed O. minuta (2n = 48, BBCC)-specific clones comprising of 105 clones (266–715 bp) from the initial library composed of 1,920 clones against O. sativa by representational difference analysis (RDA), a subtractive cloning method and validated through Southern blot hybridization. Chromosomal location of O. minuta-specific clones was identified by hybridization with the genomic DNA of eight monosomic alien additional lines (MAALs). The 37 clones were located either on chromosomes 6, 7, or 12. Different hybridization patterns between O. minuta-specific clones and wild species such as O. punctata, O. officinalis, O. rhizomatis, O. australiensis, and O. ridleyi were observed indicating conservation of the O. minuta fragments across Oryza spp. A highly repetitive clone, OmSC45 hybridized with O. minuta and O. australiensis (EE), and was found in 6,500 and 9,000 copies, respectively, suggesting an independent and exponential amplification of the fragment in both species during the evolution of Oryza. Hybridization of 105 O. minuta specific clones with BB- and CC-genome wild Oryza species resulted in the identification of 4 BB-genome-specific and 14 CC-genome-specific clones. OmSC45 was identified as a fragment of RIRE1, an LTR-retrotransposon. Furthermore this clone was introgressed from O. minuta into the advanced breeding lines of O. sativa.







Similar content being viewed by others
References
Aggarwal RK, Brar DS, Khush GS (1997) Two new genomes in the Oryza complex identified on the basis of molecular divergence analysis using total genomic DNA hybridization. Mol Gen Genet 254:1–12. doi:10.1007/s004380050384
Ammiraju JSS, Luo M, Goicoechea JL, Wang W, Kundrna D, Mueller C, Talag J, Kim H-R, Sisneros NB, Blackmon B, Fang E, Tomkins JB, Brar DS, MacKill D, McCouch S, Kurata N, Lambert G, Galbraith DW, Arumuganathan K, Rao K, Walling JG, Gill N, Yu Y, SanMiguel P, Soderlund C, Jackson S, Wing RA (2006) The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res 16:140–147. doi:10.1101/gr.3766306
Brar DS, Khush GS (2006) Cytogenetic manipulation and germplasm enhancement of rice (Oryza sativa L.). In: Jauhar PP, Singh RJ (eds) Genetic resources, chromosome engineering and crop improvement. CRC Press, Boca Raton, pp 115–158
Ge S, Sang T, Lu BR, Hong DY (1999) Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc Natl Acad Sci USA 96:14400–14405
Hu CH (1970) Cytogenetic studies of Oryza officinalis complex. III. The genomic constitution of O. punctata and O. eichingeri. Cytologia 35:304–318
Lisitsyn N, Lisitsyn NM, Wigler M (1993) Cloning the differences between two complex genomes. Science 259:946–951
Ma J, Bennetzen JL (2006) Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc Natl Acad Sci USA 103:383–388. doi:10.1073/pnas.0509810102
Martinez CP, Arumuganathan K, Kikuchi H, Earle ED (1994) Nuclear DNA content of ten rice species as determined by flow cytometry. Jpn J Genet 69:513–523
Milner JJ, Cecchini E, Dominy PJ (1995) A kinetic model for subtractive hybridization. Nucl Acids Res 23:176–187
Noma K, Nakajima R, Ohtsubo H, Ohtsubo E (1997) RIRE1, a retrotransposon from wild rice Oryza australiensis. Genes Genet Syst 72:131–140
Panaud O, Vitte C, Hivert J, Muzlak S, Talag J, Brar D, Sarr A (2002) Characterization of transposable elements in the genome of rice (Oryza sativa L.) using representational difference analysis (RDA). Mol Genet Genom 268:113–121
Park DS, Lee SK, Lee JH, Song MY, Song SY, Kwak DY, Yeo US, Jeon NS, Park SK, Yi G, Song YC, Nam MH, Ku YC, Jeon JS (2007) The identification of candidate rice genes that confer resistance to the brown planthopper (Nilaparvata lugens) through representational difference analysis. Theor Appl Genet 115:537–547. doi:10.1007/s00122-007-0587-0
Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, Kim H-R, Collura K, Brar DS, Jackson S, Wing RA, Panaud O (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16:1262–1269. doi:10.1101/gr.5290206
Toder R, Xia Y, Bausch E (1998) Interspecies comparative genome hybridization and interspecies representational difference analysis reveal gross DNA differences between humans and great apes. Chromosome Res 6:487–494. doi:10.1023/A:1009208730186
Uozu S, Ikehash H, Ohmido N, Ohtsubo H, Ohtsubo E, Fukui K (1997) Repetitive sequences: cause for variation in genome size and chromosome morphology in the genus Oryza. Plant Mol Biol 35:791–799. doi:10.1023/A:1005823124989
Vaughan DA (1994) The wild relatives of rice: a genetic resources handbook. International Rice Research Institute, Los Baños, pp 137
Wang ZY, Second G, Tanksley SD (1992) Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theor Appl Genet 83:565–581. doi:10.1007/BF00226900
Wu T, Wu R (1987) A new rice repetitive DNA shows sequence homology to both 5s RNA and tRNA. Nucl Acids Res 15:5913–5923
Yan HH, Liu GQ, Cheng ZK, Li XB, Liu GZ, Min SK, Zhu LH (2002) A genome-specific repetitive DNA sequence from Oryza eichingeri: characterization, localization, and introgression to O. sativa. Theor Appl Genet 84:136–144. doi:10.1007/s001220100766
Zhao X, Kochert G (1992) Characterization and genetic mapping of a short, highly repeated interspersed DNA sequence from rice (Oryza sativa L.). Mol Gen Genet 231:353–359
Zhao X, Wu T, Xie Y, Wu R (1989) Genome-specific repetitive sequences in the genus Oryza. Theor Appl Genet 78:201–209. doi:10.1007/BF00288800
Zoldos V, Siljak-Yakovlev S, Papes D, Sarr A, Panaud O (2001) Representational difference analysis reveals genomic differences between Q. robus and Q. suber: implications for the study of genome evolution in the genus Quercus. Mol Genet Genom 265:234–241
Acknowledgments
The senior author is thankful to the Rural Development Administration, Korea, for financial support. The authors are also grateful to Dr. Bill Hardy, senior science editor, IRRI, for editing this manuscript, to Dr. M. Ashikari for allowing to sequence 105 O. minuta-specific clones, and to K. McNally, S. Heuer, and R.A. Shim for their useful comments on it.
Author information
Authors and Affiliations
Corresponding author
Additional information
The O. minuta-specific clones are being maintained as inserts in the form of plasmids in E. coli at −80°C and are available for sharing with interested scientists. The 105 clones were sequenced and deposited in Genbank (NCBI).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shim, J., Panaud, O., Vitte, C. et al. RDA derived Oryza minuta-specific clones to probe genomic conservation across Oryza and introgression into rice (O. sativa L.). Euphytica 176, 269–279 (2010). https://doi.org/10.1007/s10681-010-0245-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-010-0245-5