Abstract
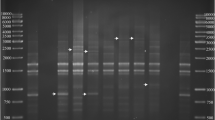
The plasmid content of 38 Iranian Erwinia amylovora strains isolated from five different plant species and geographical locations from 2005 to 2013 were evaluated. With exception to the pEA29 plasmid, isolates obtained from quince and hawthorns carry the pEU72 plasmid. RFLP analysis of PstI amplified fragment of plasmid pEA29 using HpaII revealed three known fragments in all strains. Based on the observed polymorphism in the larger one that varied between 360 to 400 bp, all Iranian strains clustered into four groups. In order to determine the exact number of short-sequence DNA repeats, 18 strains were chosen for PstI fragment sequencing. About 63 % of the strains carried a repeat number of 4. The remaining 37 % harboured five, seven and eight repeats. Sometimes a correlation was seen between the RFLP patterns and SSR grouping with respect to other factors including the host plant species, geographical region or sampling year. This study reported the genetic variation of E. amylovora strains for the first time in Iran, based on enzymatic digestion of the PstI fragment of pEA29 and evaluation of the short sequence repeat units. It seems that, the latter method may be reliable for E. amylovora strains grouping.




Similar content being viewed by others
References
Atanasova, I., Kabadjova-Hristova, P., Stefanova, K., Bogatzevska, N., & Moncheva, P. (2009). Differentiation of Erwinia amylovora strains from Bulgaria by PCR-RFLP analysis. European Journal of Plant Pathology, 124, 451–456.
Ayers, S. H., Rupp, P., & Johnson, W. T. (1919). A study of the alkali-forming bacteria in milk. United States Department of Agriculture Bulletin, 782, 1–39.
Barionovi, D., Giorgi, S., Stoeger, A. R., Ruppitsch, W., & Scortichini, M. (2006). Characterization of Erwinia amylovora strains from different host plants using repetitive sequences PCR analysis, and restriction fragment length polymorphism and short sequence DNA repeat of plasmid pEA29. Journal of Applied Microbiology, 100, 1084–1094.
Bereswill, S., Pahl, A., Bellemann, P., Zeller, W., & Geider, K. (1992). Sensitive and species-specific detection of Erwinia amylovora by PCR-analysis. Applied and Environmental Microbiology, 58, 3522–3526.
Bereswill, S., Jock, S., Bellemann, P., & Geider, K. (1998). Identification of Erwinia amylovora by growth morphology on agar containing copper sulfate and by capsule staining with lectin. Plant Disease, 82, 158–164.
Bühlmann, A., Dreo, T., Rezzonico, F., Pothier, J. F., Smits, T. H. M., Ravnikar, M., Frey, J. E., & Duffy, B. (2013). Phylogeography and population structure of the biologically invasive phytopathogen Erwinia amylovora inferred using minisatellites. Environmantal Microbiology. doi:10.1111/1462-2920.12289.
Falkenstein, H., Zeller, W., & Geider, K. (1989). The 29 kb plasmid, common in strains of Erwinia amylovora, modulates development of fire blight symptoms. Journal of General Microbiology, 135, 2643–2650.
Foster, G. C., McGhee, G. C., Jones, A. L., & Sundin, G. W. (2004). Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Applied and Environmental Microbiology, 70, 7539–7544.
Geider, K., Auling, G., Jakovljevic, V., & Volksch, B. (2009). A polyphasic approach assigns the pathogenic Erwinia strains from diseased pear trees in Japan to Erwinia pyrifoliae. Letters in Applied Microbiology, 48, 324–330.
Geier, G., & Geider, K. (1993). Characterization and influence on virulence of the levansucrase gene from the fire blight pathogen Erwinia amylovora. Physiological and Molecular Plant Pathology, 42, 387–404.
Hugh, R., & Leifson, E. (1953). The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by various gram negative bacteria. Journal of Bacteriology, 66, 24–26.
Jock, S., Jacob, T., Kim, W. S., Hildebrand, M., Vosberg, H. P., & Geider, K. (2003). Instability of short-sequence DNA repeats of pear pathogenic Erwinia strains from Japan and Erwinia amylovora fruit tree and raspberry strains. Molecular Genetics and Genomics, 268, 739–749.
Kim, W. S., & Geider, K. (1999). Analysis of variable short sequence DNA repeats on the 29 kp plasmid of Erwinia amylovora strains. European Journal of Plant Pathology, 105, 703–713.
Lecomte, P., Manceau, C., Paulin, J. P., & Keck, M. (1997). Identification by PCR analysis on plasmid pEa29 of isolates of Erwinia amylovora responsible of an outbreak in Central Europe. European Journal of Plant Pathology, 103, 91–98.
Lelliott, R. A., & Stead, D. (1987). Methods for the Diagnosis of Bacterial Diseases of Plants. Oxford: Blackwell scientific publications. 215 pp.
Llop, P., Donat, V., & Rodriguez, M. (2006). An indigenous virulent strain of Erwinia amylovora lacking the ubiquitous plasmid pEA29. Phytopathology, 96, 900–907.
Llop, P., Cabrefiga, J., Ruz, L., Montesinos, E., & Lopez, M. M. (2008). Study of virulence in wild Erwinia amylovora strains devoid of the pEA29 plasmid. Acta Horticulturae, 793, 145–148.
Llop, P., Cabrefiga, J., Smits, T. H. M., Dreo, T., Barbe, S., Pulawska, J., Bultreys, A., Blom, J., Duffy, B., Montesinos, E., & Lopez, M. M. (2011). Erwinia amylovora novel plasmid pE170: Complete sequence, biogeography, and role in aggressivenass in the fire blight phytopathogen. PloS One. doi:10.1371/journal.pone.0028651.
Llop, P., Barbe, S., & Lopez, M. M. (2012). Functions and origin of plasmids in Erwinia species that are pathogenic to or epiphytically associated with pome fruit trees. Tress, 26, 31–46.
McGhee, G. C., & Jones, A. L. (2000). Complete nucleotide sequence of ubiquitous plasmid pEA29 from Erwinia amylovora strain Ea88: gene organization and intraspecies variation. Applied and Environmental Microbiology, 66, 4897–4907.
McGhee, G. C., & Sundin, G. W. (2008). Thiamin biosynthesis and its influence on exopolysaccharide production: a new component of virulence identified on Erwinia amylovora plasmid pEA29. Acta Horticulturae, 793, 271–277.
McGhee, G. C., Schnabel, E. L., Maxson-Stein, K., Jones, B., Stromberg, V. K., et al. (2002). Relatedness of chromosomal and plasmid DNAs of Erwinia pyrifoliae and Erwinia amylovora. Applied and Environmental Microbiology, 68, 6182–6192.
McManus, P. S., & Jones, A. C. (1995). Detection of Erwinia amylovora by nested PCR and PCR-dot blot and reverse blot hybridization. Phytopathology, 85, 618–623.
Mohammadi, M. (2010). Enhanced colonization and pathogenicity of Erwinia amylovora strains transformed with the near-ubiquitous pEA29 plasmid on pear and apple. Plant Pathology, 59, 252–261.
Mohammadi, M., Moltmann, E., Zeller, W., & Geider, K. (2009). Characterization of naturally occurring Erwinia amylovora strains lacking the common plasmid pEA29 and their detection with real-time PCR. European Journal of Plant Pathology, 124, 293–302.
Niknejad Kazempour, M., Kamran, E., & Ali, B. (2006). Erwinia amylovora causing fire blight of pear in the Guilan province of Iran. Journal of Plant Pathology, 88, 113–116.
Oh, C. S., & Beer, S. V. (2005). Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiology Letters, 253, 185–192.
Palmer, E. L., Teviotdale, B. L., & Jones, A. L. (1997). A relative of the broad host range plasmid RSF1010 detected in Erwinia amylovora. Applied and Environmental Microbiology, 63, 4604–4607.
Powney, R., Smits, T. H. M., Sawbridge, T., Frey, B., Blom, J., Frey, J. E., Plummer, K. M., Beer, S. V., Luck, J., Duffy, B., & Rodoni, B. (2011). Genome sequence of an Erwinia amylovora strain with pathogenicity restricted to Rubus plants. Journal of Bacteriology, 193, 785–786.
Puławska, J., & Sobiczewski, P. (2012). Phenotypic and genetic diversity of Erwinia amylovora: the causal agent of fire blight. Trees, 26, 3–12.
Ruppitsch, W., Stöger, A., & Keck, M. (2004). Stability of short sequence repeats and their application for the characterization of Erwinia amylovora strains. FEMS Microbiology Letters, 234, 1–8.
Schaad, N. W., Jones, J. B., & Chun, W. (2001). Laboratory Guide for Identification of Plant Pathogenic Bacteria. St. Paul: APS Press. 158 pp.
Schnabel, E. L., & Jones, A. L. (1998). Instability of a pEA29 marker in Erwinia amylovora previously used for strain classification. Plant Disease, 82, 1334–1336.
Sebaihia, M., Bocsanczy, A. M., Biehl, B. S., Quail, M. A., Pema, N. T., Glasner, J. D., DeClerck, G. A., Cartinjour, S., Schneider, D. J., Bentley, S. D., Parkhill, J., & Beer, S. V. (2010). Complete genome of the plant pathogen Erwinia amylovora strain ATCC 49946. Journal of Bacteriology, 192, 2020–2021.
Smits, T. H. M., Rezzonico, F., Kamber, T., Blom, J., Goesmann, A., Frey, J. E., & Duffy, B. (2010). Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP 1430 and comparison to other Erwinia spp. Molecular Plant Microbe Interaction, 23, 384–393.
Sundin, G. W. (2007). Genomic insights into the contribution of phytopathogenic bacterial plasmids to the evolutionary history of their hosts. Annual Review of Phytopathology, 45, 129–151.
Sundin, G. W., & Murillo, J. (1999). Functional analysis of the Pseudomonas syringae rulAB determinant in tolerance to ultraviolet B (290–320 nm) radiation and distribution of rulAB among P. syringae pathovars. Environmental Microbiology, 1, 75–87.
Sundin, G. W., Jones, A. L., & Fulbright, D. W. (1989). Copper resistance in Pseudomonas syringae pv. syringae and its associated transfer in vitro and in planta with a plasmid. Phytopathology, 79, 861–865.
Taylor, R. K., Guilford, P. J., Clark, R. G., Hale, C. N., & Forster, R. L. S. (2001). Detection of Erwinia amylovora in plant material using novel polymerase chain reaction (PCR) primers. New Zealand Journal of Crop and Horticultural Science, 29, 35–43.
Thompson, J. D., Gibson, T. J., Plewniak, F., & Higgins, D. G. (1997). The CLUSTALX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 24, 4876–4882.
Van Belkum, A., Scherer, S., Van Alphen, L., & Verbrugh, H. (1998). Short-sequence DNA repeats in prokaryotic genomes. Microbiological and Molecular Biology Reviews, 62, 275–293.
Vanneste, J. L. (2000). Fire blights the disease and its causative agent, Erwinia amylovora research. Hamilton New Zealand: CABI Publishing. 358 pp.
Zhao, Y., Wang, D., Nakka, S., Sundin, G. W., & Korban, S. S. (2009). Systems level analysis of two-component signal transduction systems in Erwinia amylovora: role in virulence, regulation of amylovoran biosynthesis and swarming motility. BMC Genomics, 10, 245.
Acknowledgments
This research was financially supported by Ferdowsi University of Mashhad, Iran.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Taghdareh, G., Baghaee-Ravari, S., Moslemkhani, C. et al. Evaluation of repeat sequences on plasmid pEA29 of Erwinia amylovora from Iran. Eur J Plant Pathol 140, 735–744 (2014). https://doi.org/10.1007/s10658-014-0504-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-014-0504-8