Abstract
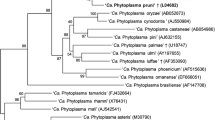
Okra plants with bunchy top disease were found to be prevalent during the period of August–October 2009 in New Delhi, India. The common symptoms observed were shortening of internodes, aggregation of leaves at the apical region, reduced leaf lamina, stem reddening, fruit bending, phyllody and stunting of plants. The disease incidence ranged from 2–60% accompanied by significant reductions in production of both flowers and seeds. Nested polymerase chain reaction targeting phytoplasma specific 16S rDNA and rp genes revealed all symptomatic plants to be positive for phytoplasma. Homology searches depicted its closest identity to phytoplasmas of 16SrI ‘Candidatus Phytoplasma asteris’, like the Sugarcane yellows and Periwinkle phyllody phytoplasmas. Profiles for 16S rDNA obtained with 10 restriction endonucleases, differed in TaqI sites for two phytoplasma isolates (BHND5 & 10) from the standard pattern of 16SrI-B subgroup, the latter was seen in the case of isolate BHND1. Restriction fragment analysis of rp genes with AluI, Tsp509I matched with patterns of the rpI-B phytoplasmas. Phylogenetic reconstruction of rp genes revealed okra bunchy top phytoplasma (BHND1) as a divergent isolate, the subsequent sequence analysis of which showed the presence of a novel BslI site. These significant differences suggest that multiple phytoplasma strains are affecting okra, one of which is a diverging lineage within the 16SrI-B group while others represent a new 16SrI subgroup not reported so far. Additionally, this is the first report of a phytoplasma associated disease in okra plants worldwide.



Similar content being viewed by others
References
Arocha, Y., Pinol, B., Picornell, B., Almeida, R., & Jones, P. (2006). First report of the 16SrII (Candidatus Phytoplasma aurantifolia) group associated with a bunchy-top disease of papaya in Cuba. Plant Pathology, 55, 821.
Chaturvedi, Y., Singh, M., Snehi, S. K., Raj, S. K., & Rao, G. P. (2009). First report of ‘Candidatus Phytoplasma asteris’ (16SrI group) associated with yellows and little leaf diseases of Hibiscus rosa-sinensis in India. New Disease Reports, 20, 25.
Deng, S., & Hiruki, C. (1991). Amplification of 16S rRNA genes from culturable and non-culturable mollicutes. Journal of Microbiological Methods, 14, 53–61.
Eckstein, B., Barbosa, J. C., Rezende, J. A. M., & Bedendo, I. P. (2011). A Sida sp. is a new host for “Candidatus Phytoplasma brasiliense” in Brazil. Plant Disease, 95, 363.
FAO (2005). http://www.fao.org/es/ess/top/commodity.html. Accessed 20 July 2011.
Khan, M. S., Singh, S. K., & Ahmad, J. (2008). Characterization and phylogeny of a phytoplasma inducing sandal spike disease in sandal (Santhalum album). Annals of Applied Biology, 153, 365–372.
Kumar, S., Singh, V., & Lakhanpaul, S. (2010a). First report of Crotalaria spectabilis fasciation associated with ‘Candidatus Phytoplasma asteris’ in India. Plant Disease, 94, 1265.
Kumar, S., Singh, V., & Lakhanpaul, S. (2010b). First report of ‘Candidatus Phytoplasma asteris’ (16SrI) associated with little leaf of cotton and luffa in India. Australasian Plant Disease Notes, 5, 117–119.
Lee, I. M., Hammond, R. W., Davis, R. E., & Gundersen, D. E. (1993). Universal amplification and analysis of pathogen 16S rDNA for classification and identification of mycoplasmalike organisms. Phytopathology, 83, 834–842.
Lee, I. M., Gundersen, D. E., Davis, R. E., & Bartoszyk, I. (1998). Revised classification scheme of phytoplasmas based on RFLP analyses of 16S rRNA and ribosomal protein gene sequences. International Journal of Systematic Bacteriology, 48, 1153–1169.
Lee, I.-M., Davis, R. E., & Gundersen-Rindal, D. E. (2000). Phytoplasma: phytopathogenic mollicutes. Annual Review of Microbiology, 54, 221–255.
Lee, I. M., Gundersen, D. E., Davis, R. E., Bottner, K. D., Marcone, C., & Seemuller, E. (2004). ‘Candidatus Phytoplasma asteris’, a novel phytoplasma taxon associated with aster yellows and related diseases. International Journal of Systematic andEvolutionary Microbiology, 54, 1037–1048.
Lee, I.-M., Zhao, Y., & Davis, R. E. (2009). Prospects for multiple gene based systems for differentiation and classification. In P. G. Weintraub & P. Jones (Eds.), Phytoplasmas. genomes, plant hosts and vectors. Wallingford: CABI. 58 pp.
Lim, P. O., & Sears, B. B. (1992). Evolutionary relationships of a plant-pathogenic mycoplasmalike organism and Acholeplasma laidlawii deduced from two ribosomal protein gene sequences. Journal of Bacteriology, 174, 2606–2611.
Marzachi, C., Coulibaly, A., Coulibaly, N., Sangare, A., Diarra, M., Gregorio, T. D., & Bosco, D. (2009). Cotton virescence phytopasma and its weed reservoir in Mali. Journal of Plant Pathology, 91, 717–721.
Montano, H. G., Davis, R. E., Dally, E. L., Hogenhout, S., Pimente, J. P., & Brioso, P. S. T. (2001). ‘Candidatus Phytoplasma brasiliense’, a new phytoplasma taxon associated with hibiscus witches’ broom disease. International Journal of Systematic and Evolutionary Microbiology, 51, 1109–1118.
Rao, G. P., Mall, S., Raj, S. K., & Snehi, S. K. (2011). Phytoplasma diseases affecting various plant species in India. Acta Phytopathologica et Entomologica Hungarica, 46, 59–99.
Saghai-Maroof, M. A., Soliman, K. M., Jorgenson, R. A., & Allard, A. W. (1984). Ribosomal spacer-length polymorphism in barley: Mendelian inheritance, chromosomal creation and population dynamics. Proceedings of the National Academy ofSciences, 81, 8014–8018.
Sambrook, J., Fritsch, E. F., & Maniatus, T. (1989). Molecular cloning: a laboratory manual (2nd ed.). New York: Cold Spring Harbor laboratory.
Seemüller, E., Schneider, B., Mäurer, R., et al. (1994). Phylogenetic classification of phytopathogenic mollicutes by sequence analysis of 16S ribosomal DNA. International Journal of Systematic Bacteriology, 44, 440–446.
Seemüller, E., Marcone, C., Lauer, U., Ragozzino, A., & Göschl, M. (1998). Current status of molecular classification of the phytoplasmas. Journal of Plant Pathology, 80, 3–26.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kumar, S., Singh, V. & Lakhanpaul, S. Molecular characterization and phylogeny of phytoplasma associated with bunchy top disease in its new host Okra (Abelmoschus esculentus) in India reveal a novel lineage within the 16SrI group. Eur J Plant Pathol 133, 371–378 (2012). https://doi.org/10.1007/s10658-011-9910-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-011-9910-3