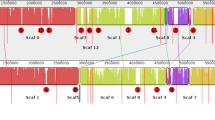
Abstract
The complete genomic sequences of several Pseudomonas spp. that inhabit the rhizosphere are now available, providing a new opportunity to advance knowledge of plant growth-promoting rhizobacteria (PGPR) through genomics. Among these is the biological control bacterium Pseudomonas fluorescens Pf-5. Nearly 6% of the 7.07 Mb genome of Pf-5 is devoted to the biosynthesis of secondary metabolites, including antibiotics toxic to soilborne fungi and Oomycetes that infect plant roots, and two siderophores involved in iron acquisition. Three orphan gene clusters, for which the encoded natural product was unknown, also were identified in the genome of Pf-5. The product synthesized from one of the orphan gene clusters was identified recently using a new ‘genomisotopic approach’, which employs a combination of genomic sequence analysis and isotope guided fractionation. Application of the genomisotopic approach to one orphan gene cluster in Pf-5 resulted in the discovery of orfamide A, founder of a new group of bioactive cyclic lipopeptides with a putative role in biological control of plant disease.



Similar content being viewed by others
Abbreviations
- CLP:
-
cyclic lipopeptide
- DAPG:
-
2,4-diacetylphloroglucinol
- GI approach:
-
genomisotopic approach
- HCN:
-
hydrogen cyanide
- NRPS:
-
non-ribosomal peptide synthetase
- PGPR:
-
Plant growth-promoting rhizobacteria
- PKS:
-
polyketide synthase
- Mcf:
-
‘makes caterpillars floppy’
References
Abbas, A., McGuire, J. E., Crowley, D., Baysse, C., Dow, M., & O’Gara, F. (2004). The putative permease PhlE of Pseudomonas fluorescens F113 has a role in 2,4-diacetylphloroglucinol resistance and in general stress tolerance. Microbiology, 150, 2443–2450.
Abbas, A., Morrissey, J. P., Marquez, P. C., Sheehan, M. M., Delany, I. R., & O’Gara, F. (2002). Characterization of interactions between the transcriptional repressor PhlF and its binding site at the phlA promoter in Pseudomonas fluorescens F113. Journal of Bacteriology, 184, 3008–3016.
Ansari, M. Z., Yadav, G., Gokhale, R. S., & Mohanty, D. (2004). NRPS-PKS: A knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Research, 32, W405–W413.
Baehler, E., Bottiglieri, M., Pechy-Tarr, M., Maurhofer, M., & Keel, C. (2005). Use of green fluorescent protein-based reporters to monitor balanced production of antifungal compounds in the biocontrol agent Pseudomonas fluorescens CHA0. Journal of Applied Microbiology, 99, 24–38.
Bangera, M. G., & Thomashow, L. S. (1999). Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. Journal of Bacteriology, 181, 3155–3163.
Blumer, C., & Haas, D. (2000). Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Archives of Microbiology, 173, 170–177.
Bottiglieri, M., & Keel, C. (2006). Characterization of PhlG, a hydrolase that specifically degrades the antifungal compound 2,4-diacetylphloroglucinol in the biocontrol agent Pseudomonas fluorescens CHA0. Applied and Environmental Microbiology, 72, 418–427.
Brodhagen, M., Paulsen, I., & Loper, J. E. (2005). Reciprocal regulation of pyoluteorin production with membrane transporter gene expression in Pseudomonas fluorescens Pf-5. Applied and Environmental Microbiology, 71, 6900–6909.
Challis, G. L., Ravel, J., & Townsend, C. A. (2000). Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chemistry & Biology, 7, 211–224.
Cipollone, R., Frangipani, E., Tiburzi, F., Imperi, F., Ascenzi, P., & Visca, P. (2007). Involvement of Pseudomonas aeruginosa rhodanese in protection from cyanide toxicity. Applied and Environmental Microbiology, 73, 390–398.
Cooper, M., Tavankar, G. R., & Williams, H. D. (2003). Regulation of expression of the cyanide-insensitive terminal oxidase in Pseudomonas aeruginosa. Microbiology, 149, 1275–1284.
Cronin D., Möenne-Loccoz, Y., Fenton, A., Dunne, C., Dowling, D. N., & O’Gara, F. (1997). Role of 2,4-diacetylphloroglucinol in the interactions of the biocontrol pseudomonad strain F113 with the potato cyst nematode Globodera rostochiensis. Applied and Environmental Microbiology, 63, 1357–1361.
Daborn, P. J., Waterfield, N., Silva, C. P., Au, C. P. Y., Sharma, S., & ffrench-Constant, R. H. (2002). A single Photorhabdus gene, makes caterpillars floppy (mcf), allows Escherichia coli to persist within and kill insects. Proceedings of the National Academy of Sciences of the United States of America, 99, 10742–10747.
Delany, I., Sheehan, M. M., Fenton, A., Bardin, S., Aarons, S., & O’Gara, F. (2000). Regulation of production of the antifungal metabolite 2,4-diacetylphloroglucinol in Pseudomonas fluorescens F113: genetic analysis of phlF as a transcriptional repressor. Microbiology, 146, 537–543.
de Bruijn, I., de Kock, M. J. D., Yang, M., de Waard, P., van Beek, T. A., & Raaijmakers, J. M. (2007). Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Molecular Microbiology, 63, 417–428.
De Souza, J. T., De Boer, M., De Waard, P., Van Beek, T. A., & Raaijmakers, J. M. (2003). Biochemical, genetic, and zoosporicidal properties of cyclic lipopeptide surfactants produced by Pseudomonas fluorescens. Applied and Environmental Microbiology, 69, 7161–7172.
Dong, C., Flecks, S., Unversucht, S., Haupt, C., van Pée, K.-H., & Naismith, J. H. (2005). Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination. Science, 309, 2216–2219.
dos Santos, V. A. P. M., Heim, S., Moore, E. R. B., Strätz, M., & Timmis, K. N. (2004). Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environmental Microbiology, 6, 1264–1286.
Feil, H., Feil, W. S., Chain, P., Larimer, F., DiBartolo, G., Copeland, A., Lykidis, A., Trong, S., Nolan, M., Goltsman, E., Thiel, J., Malfatti, S., Loper, J. E., Lapidus, A., Detter, J. C., Land, M., Richardson, P. M., Kyrpides, N. C., Ivanova, N., & Lindow, S. E. (2005). Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proceedings of the National Academy of Sciences of the United States of America, 102, 11064–11069.
Finking, R., & Marahiel, M. A. (2004). Biosynthesis of nonribosomal peptides. Annual Review of Microbiology, 58, 453–488.
Folders, J., Algra, J., Roelofs, M. S., van Loon, L. C., Tommassen, J., & Bitter, W. (2001). Characterization of Pseudomonas aeruginosa chitinase, a gradually secreted protein. Journal of Bacteriology, 183, 7044–7052.
Gross, H., Stockwell, V. O., Henkels, M. D., Nowak-Thompson, B., Loper, J. E., & Gerwick, W. H. (2007). The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chemistry & Biology, 14, 53–63.
Guenzi, E., Galli, G., Grgurina, I., Gross, D. C., & Grandi, G. (1998). Characterization of the syringomycin synthetase gene cluster. A link between prokaryotic and eukaryotic peptide synthetases. Journal of Biological Chemistry, 273, 32857–32863.
Haas, D., & Defago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nature Reviews Microbiology, 3, 307–319.
Haas, D., & Keel, C. (2003). Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annual Review of Phytopathology, 41, 117–153.
Hammer, P. E., Burd, W., Hill, D. S., Ligon, J. M., & van Pée, K. (1999). Conservation of the pyrrolnitrin biosynthetic gene cluster among six pyrrolnitrin-producing strains. FEMS Microbiology Letters, 180, 39–44.
Hammer, P. E., Hill, D. S., Lam, S. T., Van Pée, K. H., & Ligon, J. M. (1997). Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Applied and Environmental Microbiology, 63, 2147–2154.
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., & Lorito, M. (2004). Trichoderma species – opportunistic, avirulent plant symbionts. Nature Reviews Microbiology, 2, 43–56.
Helmann, J. D. (2002). The extracytoplasmic function (ECF) sigma factors. Advances in Microbial Physiology, 46, 47–110.
Howell, C. R., & Stipanovic, R. D. (1979). Control of Rhizoctonia solani in cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology, 69, 480–482.
Howell, C. R., & Stipanovic, R. D. (1980). Suppression of Pythium ultimum induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic pyoluteorin. Phytopathology, 70, 712–715.
Huang, X. Q., Yan, A., Zhang, X. H., & Xu, Y. Q. (2006). Identification and characterization of a putative ABC transporter PltHIJKN required for pyoluteorin production in Pseudomonas sp. M18. Gene, 376, 68–78.
Huang, X., Zhu, D., Ge, Y., Hu, H., Zhang, X., & Xu, Y. (2004). Identification and characterization of pltZ, a gene involved in the repression of pyoluteorin biosynthesis in Pseudomonas sp. M18. FEMS Microbiology Letters, 232, 197–202.
Keel, C., Schnider, U., Maurhofer, M., Voisard, C., Laville, J., Burger, U., Wirthner, P., Haas, D., & Défago, G. (1992). Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Molecular Plant-Microbe Interactions, 5, 4–13.
Kiil, K., Binnewies, T. T., Sicheritz-Pontén, T., Willenbrock, H., Hallin, P. F., Wassenaar, P. F., & Ussery, D. W. (2005a). Genome update: sigma factors in 240 bacterial genomes. Microbiology, 151, 3147–3150.
Kiil, K., Ferchaud, J. P., David, C., Binnewies, T. T., Wu, H., Sicheritz-Pontén, T., Willenbrock, H., & Ussery, D. W. (2005b). Genome update: distribution of two-component transduction systems in 250 bacterial genomes. Microbiology, 151, 3447–3452.
Kirner, S., Hammer, P. E., Hill, D. S., Altmann, A., Fischer, I., Weislo, L. J., Lanahan, M., van Pée, K. H., & Ligon, J. M. (1998). Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. Journal of Bacteriology, 180, 1939–1943.
Knowles, C. J. (1976). Microorganisms and cyanide. Bacteriological Reviews, 40, 652–680.
Kraus, J., & Loper, J. E. (1992). Lack of evidence for a role of antifungal metabolite production by Pseudomonas fluorescens Pf-5 in biological control of Pythium damping-off of cucumber. Phytopathology, 82, 264–271.
Kraus, J., & Loper, J. E. (1995). Characterization of a genomic region required for production of the antibiotic pyoluteorin by the biological control agent Pseudomonas fluorescens Pf-5. Applied and Environmental Microbiology, 61, 849–854.
Kuiper, I., Lagendijk, E. L., Pickford, R., Derrick, J. P., Lamers, G. E. M., Thomas-Oates, J. E., Lugtenberg, B. J. J., & Bloemberg, G. V. (2004). Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Molecular Microbiology, 51, 97–113.
Laville, J., Blumer, C., Von Schroetter, C., Gaia, V., Defago, G., Keel, C., & Haas, D. (1998). Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. Journal of Bacteriology, 180, 3187–3196.
Ligon, J. M., Hill, D. S, Hammer, P. E., Torkewitz, N. R., Hofmann, D., Kempf, H.-J., & van Pée, K.-H. (2000). Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Management Science, 56, 688–695.
Loper, J. E., & Henkels, M. D. (1999). Utilization of heterologous siderophores enhances levels of iron available to Pseudomonas putida in the rhizosphere. Applied and Environmental Microbiology, 65, 5357–5363.
Loper, J. E., Kobayashi, D. Y., & Paulsen, I. T. (2007). The genomic sequence of Pseudomonas fluorescens Pf-5: insights into biological control. Phytopathology, 97, 233–238.
Maurhofer, M., Keel, C., Haas, D., & Défago, G. (1995). Influence of plant species on disease suppression by Pseudomonas fluorescens strain CHA0 with enhanced antibiotic production. Plant Pathology, 44, 40–50.
Mavrodi, D. V., Blankenfeldt, W., & Thomashow, L. S. (2006). Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annual Review of Phytopathology, 44, 417–445.
Mavrodi, D. V., Paulsen, I. T., Ren, Q., & Loper, J. E. (2007). Genomics of Pseudomonas fluorescens Pf-5. In A. Filloux & J. L. Ramos (Eds.), Pseudomonas (Vol. V, pp. 1–28). The Netherlands: Springer Press.
Mazurier, S., Lemunier, M., Siblot, S., Mougel, C., & Lemanceau, P. (2004). Distribution and diversity of type III secretion system-like genes in saprophytic and phytopathogenic fluorescent pseudomonads. FEMS Microbiology Ecology, 49, 455–467.
Mellano, M. A., & Cooksey, D. A. (1998). Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. Journal of Bacteriology, 170, 2879–2883.
Miller, C. M., Miller, R. V., Garton-Kenny, D., Redgrave, B., Sears, J., Condron, M. M., Teplow, D. B., & Strobel, G. A. (1998). Ecomycins, unique antimycotics from Pseudomonas viridiflava. Journal of Applied Microbiology, 84, 937–944.
Nielsen, T. H., Nybroe, O., Koch, B., Hansen, M., & Sorensen, J. (2005). Genes involved in cyclic lipopeptide production are important for seed and straw colonization by Pseudomonas sp. strain DSS73. Applied and Environmental Microbiology, 71, 4112–4116.
Nielsen, T. H., Sorensen, D., Tobiasen, C., Andersen, J. B., Christophersen, C., Givskov, M., & Sorensen, J. (2002). Antibiotic and biosurfactant properties of cyclic lipopeptides produced by fluorescent Pseudomonas spp. from the sugar beet rhizosphere. Applied and Environmental Microbiology, 68, 3416–3423.
Nielsen, T. H., Thrane, C., Christophersen, C., Anthoni, U., & Sorensen, J. (2000). Structure, production characteristics and fungal antagonism of tensin – a new antifungal cyclic lipopeptide from Pseudomonas fluorescens strain 96.578. Journal of Applied Microbiology, 89, 992–1001.
Nowak-Thompson, B., Chaney, N., Wing, J. S., Gould, S. J., & Loper, J. E. (1999). Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. Journal of Bacteriology, 181, 2166–2174.
Nowak-Thompson, B., Gould, S. J., Kraus, J., & Loper, J. E. (1994). Production of 2,4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens Pf-5. Canadian Journal of Microbiology, 40, 1064–1066.
Nowak-Thompson, B., Gould, S. J., & Loper, J. E. (1997). Identification and sequence analysis of the genes encoding a polyketide synthase required for pyoluteorin biosynthesis in Pseudomonas fluorescens Pf-5. Gene, 204, 17–24.
Nybroe, O., & Sørensen, J. (2004). Production of cyclic lipopeptides by fluorescent pseudomonads. In J. L. Ramos (Ed.), Pseudomonas, Vol. 3, Biosynthesis of Macromolecules and Molecular Metabolism (pp. 147–172). New York: Kluwer Academic/Plenum Publishers.
Parret, A. H. A., Temmerman, K., & De Mot, R. (2005). Novel lectin-like bacteriocins of biocontrol strain Pseudomonas fluorescens Pf-5. Applied and Environmental Microbiology, 71, 5197–5207.
Paulsen, I. T., Press, C. M., Ravel, J., Kobayashi, D. Y., Myers, G. S., Mavrodi, D. V., DeBoy, R. T., Seshadri, R., Ren, Q., Madupu, R., Dodson, R. J., Durkin, A. S., Brinkac, L. M., Daugherty, S. C., Sullivan, S. A., Rosovitz, M. J., Gwinn, M. L., Zhou, L., Schneider, D. J., Cartinhour, S. W., Nelson, W. C., Weidman, J., Watkins, K., Tran, K., Khouri, H., Pierson, E. A., Pierson, L. S. III, Thomashow, L. S., & Loper, J. E. (2005). Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nature Biotechnology, 23, 873–878.
Pfender, W. F., Kraus, J., & Loper, J. E. (1993). A genomic region from Pseudomonas fluorescens Pf-5 required for pyrrolnitrin production and inhibition of Pyrenophora tritici-repentis in wheat straw. Phytopathology, 83, 1223–1228.
Preston, G. M., Bertrand, N., & Rainey, P. B. (2001). Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Molecular Microbiology, 41, 999–1014.
Raaijmakers, J. M., de Bruijn, I., & de Kock, M. J. (2006). Cyclic lipopeptide production by plant-associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Molecular Plant-Microbe Interactions, 19, 699–710.
Raaijmakers, J. M., Vlami, M., & de Souza, J. T. (2002). Antibiotic production by bacterial biocontrol agents. Antonie van Leeuwenhoek, 81, 537–547.
Ramette, A., Frapolli, M., Défago, G., & Moënne-Loccoz, Y. (2003). Phylogeny of HCN synthase-encoding hcnBC genes in biocontrol fluorescent pseudomonads and its relationship with host plant species and HCN synthesis ability. Molecular Plant-Microbe Interactions, 16, 525–535.
Rao, K. V., & Reddy, G. C. (1990). Synthesis and herbicidal activity of the halo analogues of pyoluteorin. Journal of Agricultural and Food Chemistry, 38, 1260–1263.
Rezzonico, F., Binder, C., Defago, G., & Moenne-Loccoz, Y. (2005). The type III secretion system of biocontrol Pseudomonas fluorescens KD targets the phytopathogenic chromista Pythium ultimum and promotes cucumber protection. Molecular Plant-Microbe Interactions, 18, 991–1001.
Rodriguez, F., & Pfender, W. F. (1997). Antibiosis and antagonism of Sclerotinia homoeocarpa and Drechslera poae by Pseudomonas fluorescens Pf-5 in vitro and in planta. Phytopathology, 87, 614–621.
Roongsawang, N., Hase, K., Haruki, M., Imanaka, T., Morikawa, M., & Kanaya, S. (2003). Cloning and characterization of the gene cluster encoding arthrofactin synthetase from Pseudomonas sp. MIS38. Chemistry & Biology, 10, 869–880.
Schnider-Keel, U., Seematter, A., Maurhofer, M., Blumer, C., Duffy, B., Gigot-Bonnefoy, C., Reimmann, C., Notz, R., Defago, G., Haas, D., & Keel, C. (2000). Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. Journal of Bacteriology, 182, 1215–1225.
Scholz-Schroeder, B. K., Soule, J. D., & Gross, D. C. (2003). The sypA, sypS, and sypC synthetase genes encode twenty-two modules involved in the nonribosomal peptide synthesis of syringopeptin by Pseudomonas syringae pv. syringae B301D. Molecular Plant-Microbe Interactions, 16, 271–280.
Sharifi-Tehrani, A., Zala, M., Natsch, A., Moenne-Loccoz, Y., & Defago, G., (1998). Biocontrol of soil-borne fungal plant diseases by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. European Journal of Plant Pathology, 104, 631–643.
Siddiqui, I. A., Haas, D., & Heeb, S. (2005). Extracellular protease of Pseudomonas fluorescens CHA0, a biocontrol factor with activity against the root-knot nematode Meloidogyne incognita. Applied and Environmental Microbiology, 71, 5646–5649.
Stachelhaus, T., Mootz, H. D., & Marahiel, M. A. (1999). The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chemistry & Biology, 6, 493–505.
Stanghellini, M. E., & Miller, R. M. (1997). Biosurfactants: their identity and potential efficacy in the biological control of zoosporic plant pathogens. Plant Disease, 81, 4–12.
Stutz, E. W., Defago, G., & Kern, H. (1986). Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology, 76, 181–185.
Thrane, C., Nielsen, T. H., Nielsen, M. N., Sorensen, J., & Olsson, S. (2000). Viscosinamide-Pseudomonas fluorescens DR54 exerts a biocontrol effect on Pythium ultimum in sugar beet rhizosphere. FEMS Microbiology Ecology, 33, 139–146.
Tripathi, R. K., & Gottlieb, D. (1969). Mechanism of action of the antifungal antibiotic pyrrolnitrin. Journal of Bacteriology, 100, 310–318.
Tsuge, K., Akiyama, T., & Shoda, M. (2001). Cloning, sequencing, and characterization of the iturin A operon. Journal of Bacteriology, 183, 6265–6273.
Voisard, C., Keel, C., Haas., D., & Défago, G. (1989). Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO Journal, 8, 351–358.
Weller, D. M., Landa, B. B., Mavrodi, O. V., Schroeder, K. L., De La Fuente, L., Blouin Bankhead, S., Allende Molar, R., Bonsall, R. F., Mavrodi, D. V., & Thomashow, L. S. (2007). Role of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in the defense of plant roots. Plant Biology, 9, 4–20.
Weller, D. M., Raaijmakers, J. M., McSpadden Gardener, B. B., & Thomashow, L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annual Review of Phytopathology, 40, 309–348.
Wenzel, S. C., & Müller, R. (2005). Formation of novel secondary metabolites by bacterial multimodular assembly lines: Deviations from textbook biosynthetic logic. Current Opinion in Chemical Biology, 9, 447–458.
Xu, G. W., & Gross, D. C. (1986). Selection of fluorescent pseudomonads antagonistic to Erwinia carotovora and suppressive of potato seed piece decay. Phytopathology, 76, 414–422.
Acknowledgements
The Pf-5 genomic sequencing project was supported by Initiative for Future Agriculture and Food Systems Grant no. 2001-52100-11329 from the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service. Research in the Loper laboratory is funded by the U.S. Department of Agriculture, Agricultural Research Service CRIS project 5358-12220-002-00D. H.G. thanks the German Research Foundation for a Research Fellowship (GR 2673/1-1).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Loper, J.E., Gross, H. Genomic analysis of antifungal metabolite production by Pseudomonas fluorescens Pf-5. Eur J Plant Pathol 119, 265–278 (2007). https://doi.org/10.1007/s10658-007-9179-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-007-9179-8