Abstract
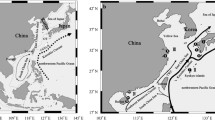
The genetic structure and demographic history of nine mudskipper Boleophthalmus pectinirostris populations on the Northwestern Pacific coast were investigated using the complete mitochondrial cytochrome b sequences. Among the 248 individuals examined, 118 haplotypes were detected. As a whole, high haplotype diversity and moderate nucleotide diversity were observed within the populations. The topology of the neighbor-joining tree of B. pectinirostris haplotypes was assigned into two closely related lineages (A and B) that appeared to have strong geographic genetic structure. Lineage A included the populations from Jiangsu, Zhejiang, Fujian, Taiwan, Guangdong, Guangxi and Vietnam while Lineage B included populations from Japan and Korea. The pairwise comparisons of F ST were all statistically significant (P <0.05) between populations while the analysis of molecular variance showed that most of the variances were found between the two lineages. Neutrality tests and mismatch distribution analyses revealed that B. pectinirostris had undergone population expansion during the late Pleistocene. Population expansion occurred approximately 170,000 years ago for Lineage A, and 80,000 years ago for Lineage B. Sea level changes during the late Pleistocene might have been responsible for this population expansion and lineage isolation.




Similar content being viewed by others
References
Avise JC (1994) Molecular markers, natural history and evolution. Chapman and Hall, New York
Billington N, Hebert PDN (1991) Mitochondrial DNA diversity in fishes and its implications for introductions. Can J Fish Aquat Sci 48(Suppl 1):80–94
Excoffier L (2004) Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol Ecol 13:853–864
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Grant WAS, Bowen BW (1998) Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. Heredity 89:415–426
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Han ZQ, Gao TX, Yanagimoto T, Sakurai Y (2008) Deep phylogeographic break among Pennahia argentata (sciaenidae, perciformes) populations in the northwestern pacific. Fish Sci 74:770–780
Hong WS, Chen SX, Zhang QY, Wang Q (2007) Reproductive ecology of mudskipper Boleophthalmus pectinirostris. Acta Oceanol Sin 26:72–81
Imbrie J, Boyle EA, Clemens SC, Duffy A, Howard WR, Kukla G, Kutzbach J, Martinson DG, McIntyre A, Mix AC, Molfino B, Morley JJ, Peterson LC, Pisias NG, Prell WL, Raymo M, EShackleton NJ, Toggweiler JR (1992) On the structure and origin of major glaciation cycles. I. Linear responses to milankovitch forcing. Paleoceanography 7:701–738
Jin CH, Zhong AH, Huang FY (2004) The research on genetic diversity of Boleophthalmus pectinirostris L. From natural population by RAPD method. Mar Sci 28:26–30 (in Chinese with English abstract)
Kanemori Y, Takegaki T, Natsukari Y (2006) Genetic population structure of the mudskipper Boleophthalmus pectinirostris inferred from mitochondrial DNA sequences. Jpn J Ichthyol 53:133–141
Kumar S, Dudley J, Nei M, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
Lambeck K, Esat TM, Potter EK (2002) Links between climate and sea levels for the past three million years. Nature 419:199–206
Liu ZZ, Yang JQ, Wang ZQ, Tang WQ (2009) Genetic structure and population history of Beleophthalmus petinirostris in Yangtze river estuary and its southern adjacent regions. Zool Res 30:1–10 (in Chinese with English abstract)
Ma C, Cheng Q, Zhang Q, Zhuang P, Zhao Y (2010) Genetic variation of Coilia ectenes (clupeiformes: engraulidae) revealed by the complete cytochrome b sequences of mitochondrial DNA. J Exp Mar Biol Ecol 385:14–19
Maggs CA, Castilho R, Foltz D, Henzler C, Jolly MT, Kelly J, Olsen J, Perez KE, Stam W, Väinölä R, Viard F, Wares J (2008) Evaluating signatures of glacial refugia for north Atlantic benthic marine taxa. Ecology 89:108–122
Massimo G, Mario LM, Vincenzo C (2009) Life style and genetic variation in teleosts: the case of pelagic (Aphia minuta) and benthic (Gobius niger) gobies (perciformes: gobiidae). Mar Biol 156:239–252
Murdy EO (1989) A taxonomic revision and cladistic analysis of the oxudurcine gobies (gobiidae: oxudercinae). Rec Aust Mus Suppl 11:1–93
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. PNAS 76:5269–5273
Perdices A, Sayanda D, Coelho MM (2005) Mitochondrial diversity of Opsariichthys bidens (teleostei, cyprinidae) in three Chinese drainages. Mol Phylogenet Evol 37:920–927
Planes S, Doherty PJ, Bernardi G (2001) Strong genetic divergence among populations of a marine fish with limited dispersal, Acanthochromis polyacanthus, within the great barrier reef and the coral Sea. Evolution 55:2263–2273
Posada D (2003) Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr Protoc Bioinform 6:1–14
Ray N, Currat M, Excoffier L (2003) Intra-deme molecular diversity in spatially expanding populations. Mol Biol Evol 20:76–86
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, New York
Slatkin M, Hudson RR (1991) Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129:555–562
Stepien CA (1999) Phylogeographical structure of the dover sole Microstomus pacificus: the larval retention hypothesis and genetic divergence along the deep continental slope of the northeastern pacific ocean. Mol Ecol 8:923–939
Tajima F (1989a) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tajima F (1989b) The effect of change in population size on DNA polymorphism. Genetics 123:597–601
Takegaki T (2008) Threatened fishes of the world: Boleophthalmus pectinirostris (Linnaeus 1758) (gobiidae). Env Biol Fish 81:373–374
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. PNAS 22:4673–4680
Wang P (1999) Response of western pacific marginal seas to glacial cycles: paleoceanographic and sedimentological features. Mar Geol 156:5–39
Yang F, He LJ, Lei GC, Zhang AB (2012) Genetic diversity and DNA barcoding of mudskipper common species along southeast coasts of china. Chin J Ecol 31:676–683 (in Chinese with English abstract)
Zhang QY, Hong W (2006) Review and prospect of mudskipper Boleophthalmus pectinirostris studies. J Xiamen Univ (Nat Sci) 45:97–108 (in Chinese with English abstract)
Acknowledgments
Professor Atsushi Ishimatsu is thanked for providing B. pectinirotris samples of Japan’s population, Kang Ning Shen for editing the manuscript, and Professor John Hodgkiss for editing the English. This work was supported by the National Natural Science Foundation of China (No. 40976095), the Key Project of Science and Technology of Fujian Province (No. 2008 N0041), Xiamen University President Research Award (No. 2013121044), and the Project of Southern Ocean Research Center of Xiamen (No.13GST001NF13).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, W., Hong, W., Chen, S. et al. Population genetic structure and demographic history of the mudskipper Boleophthalmus pectinirostris on the northwestern pacific coast. Environ Biol Fish 98, 845–856 (2015). https://doi.org/10.1007/s10641-014-0320-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10641-014-0320-1