Abstract
Given the general pattern of invasions with severe ecological consequences commonly resulting from multiple introductions of large numbers of individuals on the intercontinental scale, we explored an example of a highly successful, ecologically significant invader introduced over a short distance, possibly via minimal propagule pressure. The Sacramento pikeminnow (Ptychocheilus grandis) has been introduced to two coastal rivers in northern California where it poses a risk to threatened and endangered fishes. We assayed variation in seven microsatellite loci and one mitochondrial DNA gene to identify the source populations and estimate founder numbers for these introductions. Our analysis suggests that successful invasion of the Eel River was likely the result of a single transfer of 3–4 effective founders from nearby within the species’ native range: Clear Lake or its outflow Cache Creek. The other introduced population (Elk River), known from only seven individuals, likely represents secondary expansion from the introduced Eel River population. Our findings highlight the threat posed by close-range invaders and the ability of some fishes to rapidly invade ecologically suitable areas despite small effective founding numbers.





Similar content being viewed by others
References
Anderson EC, Slatkin M (2007) Estimation of the number of individuals founding colonized populations. Evolution 61:972–983
Baerwald MR, May B (2004) Characterization of microsatellite loci for five members of the minnow family Cyprinidae found in the Sacramento-San Joaquin Delta and its tributaries. Mol Ecol 4:385–390
Brown LR, Moyle PB (1997) Invading species in the Eel River, California: successes, failures, and relationships with native fishes. Environ Biol Fish 49:271–291
Brown JE, Stepien CA (2009) Invasion genetics of the Eurasian round goby in North America: tracing sources and spread patterns. Mol Ecol 18:64–79
Carvalho GR, Shaw PW, Hauser L, Seghers BH, Magurran AE (1996) Artificial introductions, evolutionary change and population differentiation in Trinidadian guppies (Poecilia reticulata: Poeciliidae). Biol J Linn Soc 57:219–234
Colautti RI, Grigorovich IA, MacIsaac HJ (2006) Propagule pressure: a null model for biological invasions. Biol Invasions 8(5):1023–1037
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Drake JM, Lodge DM (2006) Allee effects, propagule pressure and the probability of establishment: risk analysis for biological invasions. Biol Invasions 8:365–375
Draper NR, Smith H (1998) Applied regression analysis, 3rd edn. Wiley, New York
Duvernell DD, Aspinwall N (1995) Introgression of Luxilus cornutus mtDNA into allopatric populations of Luxilus chrysocephalus (Teleostei: Cyprinidae) in Missouri and Arkansas. Mol Ecol 4:173–182
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Res 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Friesen TA, Ward DL (1999) Management of northern pikeminnow and implications for juvenile salmonid survival in the lower Columbia and Snake Rivers. N Amer J Fish Manag 19:406–420
García-Berthou E (2007) The characteristics of invasive fishes: what has been learned so far? J Fish Biol 71(suppl D):33–55
Girard P, Angers B (2006) Characterization of microsatellite loci in longnose dace (Rhinichthys cataractae) and interspecific amplification in five other Leuciscinae species. Mol Ecol Notes 6:69–71
Harvey BC, Nakamoto RJ (1999) Diel and seasonal movements by adult Sacramento pikeminnow (Ptychocheilus grandis) in the Eel River, northwestern California. Ecol Fresh Fishes 8:209–215
Irwin DM, Kocher TD, Wilson AC (1991) Evolution of the cytochrome b gene of mammals. J Mol Evol 32:128–144
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis genetically structured populations. BMC Genet 11:94
Kalinowsi ST (2009) How well do evolutionary trees describe genetic relationships between populations? J Heredity 102:506–513
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189
Kalinowski ST, Muhlfeld CC, Guy CS, Cox B (2010) Founding population size of an aquatic invasive species. Conserv Genet 11:2049–2053
Keck BP, Near TJ (2010) Geographic and temporal aspects of mitochondrial replacement in Nothonotus darters (Teleostei: Percidae: Etheostomatinae). Evolution 64:1410–1428
Kinziger AP, Nakamoto RJ, Anderson EC, Harvey BC (2011) Small founding number and low genetic diversity in an introduced species exhibiting limited invasion success (speckled dace, Rhinichthys osculus). Ecol Evol 1:73–84
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PS, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lockwood JL, Cassey P, Blackburn T (2005) The role of propagule pressure in explaining species invasions. Trends Ecol and Evol 20:223–228
Lombaert E, Guillemaud T, Cornuet J-M, Malausa T, Facon B, Estoup A (2010) Bridgehead effect in the worldwide invasion of the biocontrol harlequin ladybird. PLoS One 5:e9743. doi:10.1371/journal.pone.0009743
Marchetti MP, Moyle PB, Levine R (2004) Invasive species profiling? Exploring the characteristics of non- native fishes across invasion stages in California. Fresh Biol 49(5):646–661
Meirmans PG, Van Tienderen PH (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Moyle PB (2002) Inland fishes of California. University of California Press, Berkeley
Muirhead JR, Gray DK, Kelly DW, Ellis SM, Heath DD, Macisaac HJ (2008) Identifying the source of species invasions: sampling intensity vs. genetic diversity. Mol Ecol 17:1020–1035
Nakamoto RJ, Harvey BC (2003) Spatial, seasonal, and size-dependent variation in the diet of Sacramento pikeminnow in the Eel River, northwestern California. California Fish and Game 89:30–45
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Nelson SM, Flickinger SA (1992) Salinity tolerance of Colorado Squawfish, Ptychocheilus lucius (Pisces: Cyprinidae). Hydrobiologia 246:165–168
Petersen JH (1994) Importance of spatial pattern in estimating predation on juvenile salmonids in the Columbia River. Trans Amer Fish Soc 123:924–930
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Development Core Team (2011) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. ISBN 3-900051-07-0
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Ricciardi A, Simberloff D (2009) Assisted colonization is not a viable conservation strategy. Trends Ecol Evol 24:248–253
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Rognon X, Guyomard R (2003) Large extent of mitochondrial DNA transfer from Oreochromis aureus to O. niloticus in West Africa. Mol Ecol 12:435–445
Roman J, Darling JA (2007) Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol Evol 22:545–564
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Ross KG, Shoemaker DD (2008) Estimation of the number of founders of an invasive pest insect population: the fire ant Solenopsis invicta in the USA. Proc R Soc B 275:2231–2240
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Schneider S, Roessli D, Excoffier L (2000) ARLEQUIN, Version 2000: a software for population genetics data analysis. Genetics and Biometry Laboratory, Department of Anthropology. University of Geneva, Geneva
Sorensen PW, Stacey NE (2004) Brief review of fish pheromones and discussion of their possible uses in the control of non-indigenous teleost fishes. NZ J Marine and Freshwater Res 38:399–417
Spencer CC, Neigel JE, Leberg PL (2000) Experimental evaluation of the usefulness of microsatellite DNA for detecting demographic bottlenecks. Mol Ecol 9:1517–1528
Stepien CA, Brown JE, Neilson ME, Tum MA (2005) Genetic diversity in invasive species in the Great Lakes versus their Eurasian source populations: insights for risk analysis. Risk Anal 25:1043–1060
Taylor CM, Hastings A (2005) Allee effects in biological invasions. Ecol Lett 8:895–908
Acknowledgments
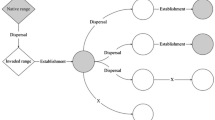
We thank the following individuals for help with field collections: Tom Kisanuki, U.S. Fish and Wildlife Service, Bill Poytress, U. S. Fish and Wildlife Service, Shawn Chase, Sonoma County Water Agency, and Rob and Daniel Gross provided invaluable assistance in providing tissue sample. Thanks to Rosemary Records for drafting Fig. 1.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kinziger, A.P., Nakamoto, R.J. & Harvey, B.C. Local-scale invasion pathways and small founder numbers in introduced Sacramento pikeminnow (Ptychocheilus grandis). Conserv Genet 15, 1–9 (2014). https://doi.org/10.1007/s10592-013-0516-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-013-0516-5