Abstract
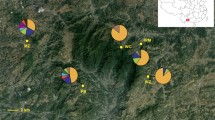
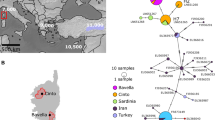
Genetic drift is expected to be the predominant evolutionary force in small, fragmented peripheral populations, which can lead to divergent allele frequencies and lowered diversity compared to the core population. Peripheral populations are not considered a high priority for conservation for this reason. However, peripheral populations may possess unique genetic variability not found elsewhere in the species’ range, and may be especially important if core populations are at extirpation risk. Here, we characterized levels and patterns of genetic diversity at microsatellites and mtDNA for the peripheral populations of Chihuahua chub in New Mexico, and compared these results to populations in Mexico including a new locality in the Rio Yaqui basin. All populations of Chihuahua chub in New Mexico were genetically depauperate as expected due to their small and peripheral status, and harbored distinct variation compared to those in Mexico. Allele and haplotype frequencies were divergent between New Mexican and Mexican populations, and mitochondrial haplotypes were not shared between them. All New Mexican populations were significantly divergent from one another suggesting little genetic exchange. New Mexican populations also exhibited relatively small genetic effective size. Chihuahua chub in New Mexico thus represent a unique component of the species’ evolutionary legacy and hence suggests high conservation value of this peripheral population. Conservation value of this population is bolstered by the fact that Chihuahua chub has more legal protection than counterparts in Mexico.



Similar content being viewed by others
References
Alò D, Turner TF (2005) Effects of habitat fragmentation on effective population size in the endangered Rio Grande silvery minnow. Conserv Biol 19:1138–1148
Baerwald MR, May B (2004) Characterization of microsatellite loci for five members of the minnow family Cyprinidae found in the Sacramento-San Joaquin Delta and its tributaries. Mol Ecol Notes 4:385–390
Baird SF, Girard C (1854) Description of new species of fishes from the United States and Canada. Third Ed. Special Publ Am Fish Soc 6:1–150
Bandelt H-J, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Beadmore JA (1983) Extinction, survival, and genetic variation. In: Schonewald-Cox CM, Chambers SM, MacBryde B, Thomas L (eds) Genetics and conservation. Benjamin Cummings Publishing, London, pp 125–151
Bowen BW (1997) Management units and evolutionary significant units in conservation. J Shellfish Res 16(1):323
Bunnell FL, Campbell RW, Squires KA (2004) Conservation priorities for peripheral species: the example of British Columbia. Can J For Res 34:2240–2247
Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK (2000) Considering evolutionary processes in conservation biology. Trends Ecol Evol 15:290–295
Dimsoski P, Toth GP, Bagley MJ (2000) Microsatellite characterization in central stoneroller Campostoma anomalum (Pisces: cyprinidae). Mol Ecol 9(12):2187–2189
Excoffier L, Laval G, Schneider S (2005) Arlequin (ver. 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fraser DF (2000) Species at the edge: the case for listing of “peripheral” species. In: Darling LM (ed) Proceedings of a Conference on the biology and management of species and habitatsat risk, 1999. Vol 1. British Columbia Ministry of Environment, Lands and Parks, Victoria, and University College of the Cariboo, Kamloops, British Columbia, Canada, pp 49–53
GerberJ AS, Tibbets CA, Dowling TE (2001) The role of introgressive hybridization in the evolution of the Gila robusta complex (Teleostei: Cyprinidae). Evolution 55:2028–2039
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Gould SJ (2002) The structure of evolutionary theory. Belknap Press of Harvard University Press, Cambridge
Hendrickson DA, Minckley WL, Miller RR, Siebert DJ, Minckley PH (1981) Fishes of the Río Yaqui Basin, Mexico and United States. J Arizona-Nevada Acad Sci 15:65–106
Hill W (1981) Estimation of effective population size from data on linkage disequilibrium. Genet Res 38:209–216
Hillis DM, Mable BK, Larson A, Davis SK, Zimmer EA (1996) Nucleic acids IV: Sequencing and cloning, 2nd edn. In: Molecular systematics. Sinauer Associates, Sunderland
Leberg PL (2002) Estimating allelic diversity: effects of sample size and bottleneck. Mol Ecol 1:2445–2449
Lesica P, Allendorf FW (1995) When are peripheral populations valuable for conservation? Conserv Biol 9:753–760
Lundberg JG (1992) The phylogeny of ictalurid catfishes: a synthesis of recent work. In: Mayden RL (ed) Systematics, historical ecology, and North American freshwater fishes. Stanford University Press, Stanford, pp 392–420
Mayr E (1954) Changes of genetic environment and evolution. In: Huxley J, Hardy AC, Ford EB (eds) Evolution as a process. Allen & Unwin, London, pp 157–180
Millar CI, Libby WJ (1991) Strategies for conserving clinal, ecotypic, and disjunct population diversity in widespread species. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York, pp 149–170
Miller RR, Chernoff B (1979) Status of populations of the endangered Chihuahua chub, Gila nigrescens, in New Mexico and Mexico. Proceedings of the Desert Fishes Council 11:74–84
Miller RR, Chernoff B (1980) Taxonomic status, ecology, and distribution of the Chihuahua chub, Gila nigrescens. Report New Mexico Department of Game and Fish, pp. 31
Miller RR, Minckley WL, Norris SM (2005) Freshwater fishes of Mexico. The University of Chicago Press, Chicago. Published in association with the Museum of Zoology, University of Michigan, pp Xxv + 490
Minckley WL, Marsh PC (2009) Inland fishes of the greater Southwest: chronicle of a vanishing biota. University of Arizona Press, Tucson
Minckley WL, Hendrickson DA, Bond CE (1986) Geography of western North American freshwater fishes: description and relationships to intracontinental tectonism. In: Hocutt CH, Wiley EO (eds) The zoogeography of North American freshwater fishes. Wiley, New York
Moritz C (1994) Defining ‘evolutionary significant units’ for conservation. Trends Ecol Evol 9:373–375
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Peakall R, Smouse P (2006) GENALEX 6: genetic analysis is Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pennock DS, Dimmick WW (1997) Critique of the evolutionarily significant unit as a definition for distinct population segments” under the US. Endangered Species Act. Conserv Biol 11:611–619
Petit RJ, El Mousadik A, Pons P (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Propst DL (1999) Threatened and endangered fishes of New Mexico. Technical Report No.1. New Mexico Department of Game and Fish, Santa Fe, New Mexico
Propst DL, Stefferud J (1994) Distribution and status of the Chihuahua chub (Teleostei: Cyprinidae: Gila nigrescens), with notes on its ecology and associated species. Southwest Nat 39:224–234
Raymond M, Rousset F (1995) GENEPOP Version 1.2: population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rogers BD (1975) Fish distribution in the Mimbres River, New Mexico. New Mexico Department of Game and Fish, pp 1–72
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Res 8:103–106
Ryman J, Laikre L (1991) Effects of supportive breeding on the genetically effective population size. Conserv Biol 5(3):325–329
Smith ML, Miller RR (1986) The evolution of the Rio Grande basin as inferred from its fish fauna. In: Hocutt CH, Wiley EO (eds) The zoogeography of North American freshwater fishes. Wiley, New York, pp 457–485
Stefferud JA, Propst DL (1996) Fish fauna of the Bavicora Basin, Chihuahua, Mexico. The Southwestern Naturalist 41:446–450
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Turner TF, Dowling DE, Broughton RE, Gold JR (2004) Variable microsatellite markers amplify across divergent lineages of cyprinid fishes (subfamily Leusicinae). Conserv Genet 5:279–381
U.S. Fish and Wildlife Service (1983) Endangered and threatened wildlife and plants: threatened status of Gila nigrescens (Chihuahua chub). Fed Reg 48:46053–46057
U.S. Fish and Wildlife Service (2010) Chihuahua chub (Gila nigrescens) five-year review: summary and evaluation. Albuquerque Office, p 23
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Waples RS (1998) Evolutionarily significant units, distinct population segments, and the Endangered Species Act: reply to Pennock and Dimmick. Conserv Biol 12:718–721
Waples RS (2006) A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv Genet 7(2):167–184
Waples RS, Do C (2008) LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Res 8:753–756
Waples RS, Do C (2009) Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3:1358–1370
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Acknowledgments
Sincere thanks are extended to Angela James (U.S. Fish and Wildlife Service) for sample collection. We are also grateful to Thomas E. Dowling and Richard L. Mayden who provided samples from Mexico, and to Paul Moreno who granted permission to sample and retain specimens from his property at Moreno Spring. Likewise, The Nature Conservancy and the New Mexico Department of Game and Fish granted permission to sample adjacent to their properties in the Mimbres River. Samples were also provided by Dexter National Fish Hatchery and Technology Center. We are extremely grateful to Alejandro Garza (ProNatura, MX) and Tyler Pilger for assistance in producing maps for this paper. Krista Heideman provided information about microsatellite primer optimization, Thien Le and Samantha Sanchez provided laboratory assistance. Funding was provided by the N.M. Department of Game and Fish Share with Wildlife Program (to MJO and TFT). New Mexico samples were collected under NM Game and Fish permit numbers: 3015 and 1776. IACUC protocol number 10-100492-MCC.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Osborne, M., Sharp, A., Monzingo, J. et al. Genetic analysis suggests high conservation value of peripheral populations of Chihuahau chub (Gila nigrescens). Conserv Genet 13, 1317–1328 (2012). https://doi.org/10.1007/s10592-012-0374-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-012-0374-6