Abstract
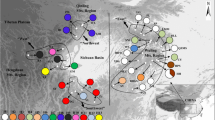
Cinnamomum kanehirae Hayata (Lauraceae), the most valuable subtropical and temperate broadleaf timber tree in Taiwan, is rapidly disappearing from the wild. Taking advantage of a scion garden established by the Taiwan Forestry Research Institute, we examined patterns of chloroplast DNA (cpDNA) variations in 19 populations including 94 individuals. By sequencing two cpDNA fragments using universal primers (the trnL-trnF and petG-trnP intergenic spacers), we found eight polymorphic sites, six haplotypes, and extremely low nucleotide diversity (π = 0.00016) from 792 bp aligned sequences. The ancestral haplotype is widely distributed. Among the populations studied, three separated populations, at Yungfeng, Fuli, and Tahu have high nucleotide diversity. No phylogeographical structures of haplotypes were revealed because the tests of NST−GST for populations did not differ from zero in any situations; a ‘star-like’ genealogy is characteristic when all haplotypes rapidly coalesce and is a general outcome of population expansion. The neutrality test also suggested demographic expansion. The genetic divergence and diversity analyses suggested that two potential refugia existed during the last glaciation with a major one located in southeastern Taiwan and a minor one located in Tahu in north-central Taiwan in the Hsuehshan Range, west of the Central Mountain Range.




Similar content being viewed by others
References
Avise JC (2000) Phylogeography: the history and formation of species. Harvard University Press, Cambridge
Bonnet E, Van De Peer Y (2002) ZT: a software tool for simple and partial Mantel tests. J Stat Softw 7:1–12
Burg TM, Lomax J, Almond R, Brooke MD, Amos W (2003) Unravelling dispersal patterns in an expanding population of a highly mobile seabird, the northern fulmar (Fulmarus glacialis). Proc R Soc Lond Ser B Biol Sci 270:979–984. doi:10.1098/rspb.2002.2322
Cheng YP, Hwang SY, Lin TP (2005) Potential refugia in Taiwan revealed by the phylogeographical study of Castanopsis carlessi Hayata (Fagaceae). Mol Ecol 14:2075–2085. doi:10.1111/j.1365-294X.2005.02567.x
Chung JD, Lin TP, Tan YC, Lin MY, Hwang SY (2004) Genetic diversity and biogeography of Cunninghamia konishii (Cupressaceae), an island species in Taiwan: a comparison with Cunninghamia lanceolata, a mainland species in China. Mol Phylogen Evol 33:791–801. doi:10.1016/j.ympev.2004.08.011
Clegg MT, Zurawski G (1992) Chloroplast DNA and the study of plant phylogeny: present status and future prospects. In: Soltis PS, Soltis DE, Doyle JJ (eds) Molecular systematics of plants. Chapman and Hall, New York, pp 1–13
Comes HP, Kadereit JW (1998) The effect of quaternary climatic changes on plant distribution and evolution. Trends Plant Sci 3:432–438. doi:10.1016/S1360-1385(98)01327-2
Comps B, Gomory D, Letouzey J, Thiebaut B, Petit RJ (2001) Diverging trends between heterozygosity and allelic richness during postglacial colonization in the European beech. Genetics 157:389–397
Corriveau JL, Coleman AW (1988) Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am J Bot 75:1443–1448. doi:10.2307/2444695
Csaikl UM, Burg K, Fineschi S, Konig AO, Matyas G, Petit RJ (2002) Chloroplast DNA variation of white oaks in the Alpine region. For Ecol Manage 156:131–145
Doyle JJ, Doyle JL (1990) Isolation plant DNA from fresh tissue. Focus 12:13–15
Dumolin-Lapègue S, Demesure B, Fineschi S, Lecorre V, Petit RJ (1997) Phylogeographic structure of white oaks throughout the European continent. Genetics 146:1475–1487
Ferris C, King RA, Vainola R, Hewitt GM (1998) Chloroplast DNA recognizes three refugial sources of European oaks and suggests independent eastern and western immigrations to Finland. Heredity 80:584–593. doi:10.1046/j.1365-2540.1998.00342.x
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Fujita Y (1967) Classification and phylogeny of the genus Cinnamomum viewed from the constituents of essential oils. Bot Mag Tokyo 80:261–271
Grant PP, Grant BR (1997) Genetics and the origin of bird species. Proc Natl Acad Sci USA 94:7768–7775. doi:10.1073/pnas.94.15.7768
Grivet D, Petit RJ (2002) Phylogeography of the common ivy (Hedera sp.) in Europe: genetic differentiation through space and time. Mol Ecol 11:1351–1362. doi:10.1046/j.1365-294X.2002.01522.x
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. In: Adams WT, Strauss SH, Copes DL, Griffin AR (eds) Population genetics of forest trees. Kluwer, Dordrecht, pp 95–124
Hare MP (2001) Prospects for nuclear gene phylogeography. Trends Ecol Evol 16:700–706. doi:10.1016/S0169-5347(01)02326-6
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc Lond 58:247–276
Hewitt GM (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913. doi:10.1038/35016000
Hsieh CF, Shen CF, Yang KC (1994) Introduction to the flora of Taiwan 3. Floristics, phytogeography, and vegetation. In: Flora of Taiwan, vol 1, 2nd edn. Editorial Committee of the Flora of Taiwan, Taipei, pp 7–18
Huang SSF, Hwang SY, Lin TP (2002) Spatial pattern of chloroplast DNA variation of Cyclobalanopsis glauca in Taiwan and East Asia. Mol Ecol 11:2349–2358. doi:10.1046/j.1365-294X.2002.01624.x
Huang SF, Hwang SY, Wang JC, Lin TP (2004) Phylogeography of Trochodendron aralioides (Trochodendraceae) in Taiwan and its adjacent areas. J Biogeogr 31:1251–1259. doi:10.1111/j.1365-2699.2004.01082.x
Huntley B, Birks HJB (1983) An atlas of past and present Pollen Maps for Europe, 0–13000 years ago. Cambridge University Press, UK
Hwang SY, Lin TP, Ma CS, Lin CL, Chung JD, Yang JC (2003) Postglacial population growth of Cunninghamia konishii (Cupressaceae) inferred from phylogeographical and mismatch analysis of chloroplast DNA variation. Mol Ecol 12:2689–2695. doi:10.1046/j.1365-294X.2003.01935.x
Johnson KP, Adler FR, Cherry JL (2000) Genetic and phylogenetic consequences of island biogeography. Evol Int J Org Evol 54:387–396
Lee JK (2003) Phylogeography of the red-bellied tree squirrel Callosciurus erythraeus of Taiwan and reexamination of its subspecific status. Master thesis (in Chinese with English summary), Tunghai University, Hualien
Lee YJ, Hwang SY, Ho KC, Lin TP (2006) Source populations of Quercus glauca in the last glacial age in Taiwan revealed by nuclear microsatellite markers. J Hered 97:261–269. doi:10.1093/jhered/esj030
Li WH (1997) Molecular evolution. Sinauer Associates, Sunderland
Lin TP (1993) Cinnamomum kanehirae Hay. and Cinnamomum micranthum (Hay.) Hay. (in Chinese with English summary). Bull Taiwan For Res Inst 8:11–20
Lin TP (2001) Allozyme variations in Michelia formosana (Kanehira) Masamune (Magnoliaceae), and the inference of glacial refugium in Taiwan. Theor Appl Genet 102:450–457. doi:10.1007/s001220051666
Lin TP, Cheng YP, Huang SG (1997) Allozyme variation in four geographic areas of Cinnamomum kanehirae. J Hered 88:433–438
Matyas G, Sperisen C (2001) Chloroplast DNA polymorphism provide evidence for postglacial re-colonisation of oaks (Quercus spp.) across the Swiss Alps. Theor Appl Genet 102:198–202
Moritz C, Faith DP (1998) Comparative phylogeography and the identification of genetically divergent areas for conservation. Mol Ecol 7:419–429. doi:10.1046/j.1365-294x.1998.00317.x
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Petit RJ, Kremar A, Wagner DB (1993) Geographic structure of chloroplast DNA polymorphisms in European oaks. Theor Appl Genet 87:122–128. doi:10.1007/BF00223755
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic marker. Conserv Biol 12:844–855. doi:10.1046/j.1523-1739.1998.96489.x
Petit RJ, Aguinagalde I, De Beaulieu JL, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, Monhanty A, Muller-Starck G, Demesure-Musch B, Palme A, Martin JP, Rendell S, Vendramin GG (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563–1565. doi:10.1126/science.1083264
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Rozas J, Sánchez-Delbarrio JC, Messegyer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497. doi:10.1093/bioinformatics/btg359
Schneider S, Roessli D, Excoffier L (2000) Arlequin ver. 2.0: a software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva
Shen CF (1997) The biogeography of Taiwan: 2. Some preliminary thoughts and studies (in Chinese with English summary). Annu Rep Taiwan Mus 40:361–450
Shih FL, Cheng YP, Hwang SY, Lin TP (2006) Partial concordance between nuclear and organelle DNA in revealing the genetic divergence among Quercus glauca (Fagaceae) populations in Taiwan. Int J Plant Sci 167:863–872. doi:10.1086/504923
Shih FL, Hwang SY, Cheng YP, Lee PF, Lin TP (2007) Uniform genetic diversity, low differentiation, and neutral evolution characterize contemporary refuge populations of Taiwan fir (Abies kawakamii, Pinaceae). Am J Bot 94:194–202. doi:10.3732/ajb.94.2.194
Taberlet P, Fumagalli L, Wust-Saucy AG, Cossons JF (1998) Comparative phylogeography and postglacial colonization routes in European. Mol Ecol 7:453–464. doi:10.1046/j.1365-294x.1998.00289.x
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619–633
Thompson JD, Higgins DG, Gibon TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi:10.1093/nar/22.22.4673
Toda M, Nishida M, Matsui M, Lue KY, Ota H (1998) Genetic variation among populations of the Indian rice frog, Rana limnocharis (Amphibia: Anura) within Taiwan: revealed by allozyme data. Herpetologica 54:78–82
Tsukada M (1966) Late Pleistocene vegetation and climate in Taiwan (Formosa). Proc Natl Acad Sci USA 55:543–584. doi:10.1073/pnas.55.3.543
Uka S (1927) Report on the survey of Taiwan camphora trees. Monopoly Bureau, Formosan Government, Taipei
Wang WP, Hwang CY, Lin TP, Hwang SY (2003) Historical biogeography and phylogenetic relationships of the genus Chamaecyparis (Cupressaceae) inferred from chloroplast DNA polymorphism. Plant Syst Evol 241:13–28. doi:10.1007/s00606-003-0031-0
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondria, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84:9054–9058. doi:10.1073/pnas.84.24.9054
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Wu SH, Hwang CY, Lin TP, Chung JD, Cheng YP, Hwang SY (2006) Contrasting phylogeographic patterns of two closely related species, Machilus thunbergii and Machilus kusanoi (Lauraceae), in Taiwan. J Biogeogr 33:936–947. doi:10.1111/j.1365-2699.2006.01431.x
Yeh WB, Chang YL, Lin CH, Wu FS, Yang JT (2004) Genetic differentiation of Loxoblemmus appendicularis complex (Orthoptera: Gryllidae): speciation through vicariant and glaciation events. Ann Entomol Soc Am 97:613–623. doi:10.1603/0013-8746(2004)097[0613:GDOLAC]2.0.CO;2
Zanetto A, Kremer A (1995) Geographical structure of gene diversity in Quercus petraea (Matt.) Liebl. I. Monolocus patterns of variation. Heredity 75:506–517. doi:10.1038/hdy.1995.167
Acknowledgments
This investigation was funded by the Council of Agriculture (grant number 95AS-12.3.1-FB-e1), Executive Yuan, Taiwan to Shih-Ying Hwang. The authors acknowledge the assistance of the Taiwan Forestry Research Institute in providing the materials of Cinnamomum kanehirae growing in a scion garden at the Liukuei Research Center.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Kuo, DC., Lin, CC., Ho, KC. et al. Two genetic divergence centers revealed by chloroplastic DNA variation in populations of Cinnamomum kanehirae Hay. Conserv Genet 11, 803–812 (2010). https://doi.org/10.1007/s10592-009-9901-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-009-9901-5