Abstract
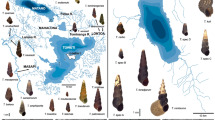
To evaluate the genetic diversity of a mangrove species and clarify the genetic structure of its populations, we studied nucleotide polymorphism in two DNA regions of Bruguiera gymnorhiza collected from the southern islands of Japan, Thailand, Malaysia, Indonesia, Micronesia, and India. The two DNA sequences were the chloroplast (cp) intergenic spacer between trnL and trnF genes (ca. 300 bp), and a part (ca. 550 bp) of the nuclear gene coding for glyceraldehyde-3-phosphate dehydrogenase (GapCp). Little polymorphism was found within each of the three geographical regions, Pacific Ocean, Bay of Bengal and Arabian Sea. Throughout the vast regions east of the Malay peninsula including Indonesia, Thailand, Micronesia and the southern islands of Japan (Pacific Ocean), essentially only one haplotype (apart from variation in number of a T repeat) was present. A second haplotype was present on the western coast of Malay Peninsula and the eastern coast of India (Bay of Bengal). On the southwest of Malay Peninsula both of these haplotypes were present. Finally a third haplotype was found only on the western coast of India (Arabian Sea). When taken over all geographic populations, total nucleotide variation within the species was large (μ = 0.006, average of the two genes). Our results are consistent with the hypothesis that this low genetic diversity within any local population and differentiation between the different oceans or regions are caused by very low gene flow between each of the different oceans coupled with frequent fluctuation of population sizes due to the change in sea level. The significance of these results is discussed from evolutionary point of the mangrove forests.




Similar content being viewed by others
References
Abeysinghe PD, Triest L, De-Greef B et al (1999) Genetic differentiation between Bruguiera gymnorhiza and B. sexangula in Sri Lanka. Hydrobiologia 5:11–16. doi:10.1023/A:1003899028558
Abeysinghe PD, Triest L, De-Greef B et al (2000) Genetic and geographic variation of the mangrove tree Bruguiera in Sri Lanka. Aquat Bot 67:131–141. doi:10.1016/S0304-3770(99)00096-0
Aksornkoae S, Maxwell G, Havanond S, Panichsuko S (1992) Plants in mangroves. Chalongrat Co. Ltd, Bangkok
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Duke NC, Benzie JAH, Goodall JA, Ballment ER (1998) Genetic structure and evolution of species in the mangrove genus Avicennia (Avicenniaceae) in the Indo-West Pacific. Evol Int J Org Evol 52:1612–1626. doi:10.2307/2411335
Elmqvist T, Cox PA (1996) The evolution of vivipary in flowering plants. Oikos 77:3–9. doi:10.2307/3545579
Fleminger A (1985) The Pleistocene equatorial barrier between the Indian and Pacific Oceans and a likely cause for Wallace’s line. UNESCO, Technical Report No. 49
Fujii N, Ueda K, Watano Y, Shimizu T (1995) Intraspecific sequence variation in chloroplast DNA of Primula cuneifolia Ledeb. (Primulaceae). J Phytogeogr Taxon 43:15–24
Fujimoto K (1997) Mangrove habitat evolution related to Holocene sea-level changes on Pacific islands. Tropics 6:203–213
Fujimoto K, Miyagi T, Kikuchi T, Kawana T (1996) Mangrove habitat formation and response to Holocene sea-level changes on Kosrae Island, Micronesia. Mangroves Marshes 1:47–57. doi:10.1023/A:1025994128221
Ge XJ, Sun M (1999) Reproductive biology and genetic diversity of a cryptoviviparous mangrove Aegiceras corniculatum (Myrsinaceae) using allozyme and intersimple sequence repeat (ISSR) analysis. Mol Ecol 8:2061–2069. doi:10.1046/j.1365-294x.1999.00821.x
Ge XJ, Sun M (2001) Population geneic structure of Ceriops tagal (Rhizophoraceae) in Thailand and China. Wetlands Ecol Manag 9:213–219. doi:10.1023/A:1011156707160
Gielly L, Taberlet P (1994) The use of chloroplast DNA to resolve plant phylogenies: Non-coding versus rbcL sequences. Mol Biol Evol 11:769–777
Hamrick J, Godt M (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc Lond B Biol Sci 351:1292–1298. doi:10.1098/rstb.1996.0112
Hatcher B, Johannes R, Robertson A (1989) Review of research relevant to conservation of shallow tropical marine ecosystems. Oceanogr Mar Biol An Annu Rev 27:337–414
Ishiyama H, Inomata N, Yamazaki T, Ab Shukor N, Szmidt AE (2006) Demographic history and interspecific hybridization of four Shorea species (Dipterocarpaceae) from Peninsula Malaysia inferred from nucleotide polymorphism in cuclear gene regions. Can J For Res 38:996–1007. doi:10.1139/X07-218
Kajita T, Kamiya K, Nakamura K et al (1998) Molecular phylogeny of Dipterocarpaceae in Southeast Asia based on nucleotide sequence of matK, trnL intron and trnL–trnF intergenic spacer region in chloroplast DNA. Mol Phylogenet Evol 10:202–209. doi:10.1006/mpev.1998.0516
Kondo K, Nakamura T, Tsuruda K et al (1987) Pollination in Bruguiera gymnorhiza and Rhizophora mucronata (Rhizophoraceae) in Ishigaki Island, the Ryukyu Islands, Japan. Biotropica 19:377–380. doi:10.2307/2388639
Lakshmi M, Rajalakshmi S, Parani M et al (1997) Molecular phylogeny of mangroves. 1. Use of molecular markers in assessing the intraspecific genetic variability in the mangrove species Acanthus ilicifolius Linn (Acanthaceae). Theor Appl Genet 94:1121–1127. doi:10.1007/s001220050525
Lavery S, Moritz C, Fielder D (1995) Changing patterns of population structure and gene flow at different spatial scales in Birgus latro (the coconut crab). Heredity 74:531–541. doi:10.1038/hdy.1995.75
Lavery S, Moritz C, Fielder D (1996) Indo-Pacific population structure and evolutionary history of the coconut crab Birgus latro. Mol Ecol 5:557–570. doi:10.1111/j.1365-294X.1996.tb00347.x
Lee T, Lawver L (1995) Cenozoic plate reconstruction of Southeast Asia. Tectonophysics 251:85–138. doi:10.1016/0040-1951(95)00023-2
Maguire T, Saenger P, Baverstock P, Henry R (2000) Microsatellite analysis of genetic structure in the mangrove species Avicennia marina (Forsk.) Vierh. (Avicenniaceae). Mol Ecol 9:1853–1862. doi:10.1046/j.1365-294x.2000.01089.x
Martin W, Lydiate D, Brinkmann H et al (1993) Molecular phylogenies in angiosperm evolution. Mol Biol Evol 10:140–162
Murray V (1989) Improved double strand DNA sequencing using the linear polymerase chain reaction. Nucleic Acids Res 17:251–257
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Noske R (1993) Bruguiera hainesii: another bird-pollinated mangrove? Biotropica 25:481–483. doi:10.2307/2388873
Parani M, Lakshmi M, Elango S et al (1997) Molecular phylogeny of mangroves II. Intra- and inter-specific variation in Avicennia revealed by RAPD and RFLP markers. Genome 40:487–495. doi:10.1139/g97-065
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497. doi:10.1093/bioinformatics/btg359
Shin MC, Heinrich P, Goodman HM (1991) Cloning and chromosomal mapping of nuclear genes encoding chloroplast and cytosolic glycerylaldehyde-3-phosphate-dehydrogenase from Arabidopsis thaliana. Gene 104:133–138. doi:10.1016/0378-1119(91)90242-4
Sugaya T, Yoshimaru H, Takeuchi T, Katsuta M, Fujimoto K (2003) Development and polymorphism of simple sequence repeat DNA markers for Bruguiera gymnorhiza (L.) Lamk. Mol Ecol Notes 3:88–90. doi:10.1046/j.1471-8286.2003.00360.x
Sun M, Wong KC, Lee JSY (1998) Reproductive biology and population genetic structure of Kandelia candel (Rhizophoraceae), a viviparous mangrove species. Am J Bot 85:1631–1637. doi:10.2307/2446492
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109. doi:10.1007/BF00037152
Takeuchi T, Sugaya T, Kanazashi A et al (2001) Genetic diversity of Kandelia candel and Bruguiera gymnorhiza in the southwest islands, Japan. J For Res 6:157–162. doi:10.1007/BF02767087
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi:10.1093/molbev/msm092. Publication PDF at http://www.kumarlab.net/publications
Tomlinson PB (1986) The botany of mangroves. Cambridge University Press, Cambridge
Tomlinson PB, Primack RB, Bunt JS (1979) Preliminary observations on floral biology in mangrove Rhizophoraceae. Biotropica 11:256. doi:10.2307/2387918
Umali R, Zamora P, Gatera R, et al (1987) Mangrove of Asia and the Pacific: status and management. Technical Report of the UNDP/UNESCO Research and Training Pilot Programme on mangrove ecosystems in Asia and the Pacific (RAS 79(002)
Wang XR, Tsumura Y, Yoshimaru H et al (1999) Phylogenetic relationships of Eurasian pines (Pinus, Pinaceae) based on chloroplast rbcL, matK, rpl20–rps18 spacer, and trnV intron sequences. Am J Bot 86:1742–1753. doi:10.2307/2656672
Williams S, Benzie J (1996) Genetic uniformity of widely separated populations of the coral reef fish Linckia laevigata from the East Indian and West Pacific Oceans, revealed by allozyme electrophoresis. Mar Biol (Berl) 126:99–107. doi:10.1007/BF00571381
Williams S, Benzie J (1997) Indo-West Pacific patterns of genetic differentiation in the high-dispersal starfish Linckia laevigata. Mol Ecol 6:559–573. doi:10.1046/j.1365-294X.1997.00221.x
Williams S, Benzie J (1998) Evidence of biogeographic break between populations of a high dispersal starfish: congruent regions within the Indo-West Pacific defined by color morphs, mtDNA, and allozyme data. Evol Int J Org Evol 52:87–99. doi:10.2307/2410923
Wolfe KH, Li W-H, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84:9054–9058. doi:10.1073/pnas.84.24.9054
Wright S (1943) Isolation by distance. Genetics 28:114–138
Acknowledgments
We thank Dr. T. Miyagi, Tohoku Gakuin University, Dr. T. Kikuchi, Gifu University, Dr. Y. Mochida, Yokohama National University and other members of the mangrove research group with whom we worked in Thailand and Malaysia in 1996 for providing opportunity for sampling in the mangrove forests and helping with identification. Thanks are also to Dr. K. Ogino, Ehime University, Dr. Mahani Mansor Clyde, Universiti Kebangsaan Malaysia, Dr. Mashhor Mansor, Universiti Sains Malaysia, Dr. M. Lokman Husain, Universiti Kolej Terengganu and many other members of laboratories which we visited in Malaysia in 1997 for providing opportunities for sampling and a place for DNA extraction, and helping with sampling and DNA extraction. We also thank Dr. K. Harada, Dr. E. Nitasaka for help in sampling of the material. Discussion with Dr. A. Szmidt of Kyushu University was helpful. We also thank Dr. Richard Frankham of Macquarie University and Dr. Peter Saenger of Southrn Cross University for careful reading of the manuscript and valuable suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Minobe, S., Fukui, S., Saiki, R. et al. Highly differentiated population structure of a Mangrove species, Bruguiera gymnorhiza (Rhizophoraceae) revealed by one nuclear GapCp and one chloroplast intergenic spacer trnF–trnL . Conserv Genet 11, 301–310 (2010). https://doi.org/10.1007/s10592-009-9806-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-009-9806-3