Abstract
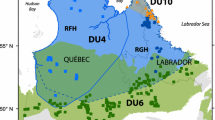
Feral pigs (Sus scrofa) are an invasive species in Australia. Their negative impact on conservation values has been demonstrated, and they are controlled in many areas in the rangelands of Australia. However, they are usually controlled over small, often ad hoc management units (MUs), and previous research has revealed that these MUs can be inadequate. Understanding feral pig population structuring can aid in the design of appropriate MUs. This study documents an approach to improving MUs for feral pig control in the rangelands of Australia. Feral pigs from a 500,000 km2 region were genotyped with 13 polymorphic markers. Genetic analyses were used to identify population structure. Identified sub-populations were then related to geographical and environmental gradients with geographical information systems, regression analysis and with canonical correspondence analysis. Five sub-populations were identified. These were moderately differentiated, with relatively high-migration rates. Two sub-populations in drier, lower elevation areas overlapped, due to extensive migration, probably along the large, inland rivers and flood plains. Sub-populations in higher rainfall environments appeared less likely to migrate. Sub-population differentiation was also dependant on distance, indicating isolation by distance was present. A case study applying an adaptive MU to a previously controlled area is presented. Generally, however, MUs for feral pig control for natural resource protection and endemic disease eradication in the rangelands should take into account geographical size, but also geographic features, especially major rivers in low-rainfall areas.






Similar content being viewed by others
References
Alexander LJ, Rohrer GA, Beattie CW (1996) Cloning and characterisation of 414 polymorphic porcine microsatellites. Anim Genet 27:137–148
Barton NH, Slatkin M (1986) A quasi-equilibrium theory of the distribution of rare alleles in a subdivided population. Heredity 56:409–415
Bomford M, O’Brien P (1995) Eradication of Australia’s vertebrate pests: a feasibility study. In: Grigg GC, Hale PT, Luney D (eds) Conservation through sustainable use of wildlife. Centre for conservation biology. The University of Queensland, Queensland, pp 243–250
Braysher M (1993) Managing vertebrate pests: principles and strategies. Bureau of Resource Sciences, Australian Government Publishing Service, Canberra
Choquenot D, McIlroy J, Korn T (1996) Managing vertebrate pests: feral pigs. Bureau of Resource Sciences, Australian Government Publishing Service, Canberra, p 163
Cockerham CC (1973) Analyses of gene frequencies. Genetics 74:679–700
Cowled BD, Lapidge SJ, Hampton JO, Spencer PBS (2006a) A cautionary finding for applied pest management: measuring the demographic and genetic effects of pest control in a highly persecuted feral pig population. J Wildl Manage 70(6):1690–1697
Cowled BD, Gifford E, Smith M, Staples L, Lapidge SJ (2006b) Efficacy of manufactured PIGOUT® baits for localised control of feral pigs in the semi-arid Queensland rangelands. Wildl Res 33:427–437
Department of Environment and Heritage (2004) Threat abatement plan for predation, habitat degradation, competition and disease transmission by feral pigs. Australian Department of Environment and Heritage, Canberra, Australia
ESRI (2006) ARCGIS & ARC/INFO User’s manual, Supplemented by ARCGIS ARC/INFO Online help. Environmental Systems Research Institute, Redlands, CA, USA
Freeland JR (2005) Molecular ecology. Wiley. Chichester, England
Goudet J (2002) FSTAT (Version 2.9.3.2): A program to estimate and test gene diversities and fixation indices. Available from http://www.unil.ch/izea/softwares/fstat.html
Hampton JO, Spencer PBS, Alpers D, Twigg L, Woolnough A, Doust J, Higgs T, Pluske J (2004) Molecular techniques, wildlife management and the importance of genetic populations structure and dispersal: a case study with feral pigs. J Appl Ecol 41:735–743
Hone J (2002) Feral pigs in Namadgi National Park: Dynamics, impacts and management. Biol Conserv 105: 231–242
Hone J, Pech R (1990) Disease surveillance in wildlife with emphasis on detecting foot-and-mouth disease in feral pigs. J Environ Manage 31:173–184
Hone J, O’Grady J, Pedersen H (1980) Decisions in the control of feral pig damage. AG Bulletin 5, Department of Agriculture, NSW
Laurence WF, Harrington GN (1997) Ecological associations of feeding sites of feral pigs in the Queensland wet tropics. Wildl Res 24:579–590
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol 18:189–197
Mantel N (1967) The detection of disease clustering and a generalised regression approach. Cancer Res 27:209–220
McCune B (1997) Influence of noisy environmental data on canonical correspondence analysis. Ecology 78:2617–2623
McKnight T (1976) Friendly vermin: a survey of feral livestock in Australia. Univ Calif Publ Geogr 21:39–55
Miller B, Mullette K (1985) Rehabilitation of an endangered Australian bird: the Lord Howe Island woodhen, Tricholimnas sybvestris. Biol Conserv 34:55–95
Mitchell J (2000) Ecology and management of feral pigs in tropical rainforest. Ph.D. thesis. James Cook University of North Queensland, Townsville
Ohta T, Kimura M (1973) A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet Res Camb 22:201–204
Palmer MW (1993) Putting things in even better order: the advantage of canonical correspondence analysis. Ecology 74:2215–2230
Palsbøll PJ, Berubé M, Allendorf FW (2007) Identification of management units using population genetic data. Trends Ecol Evol 22(1):11–16
Peakall R, Smouse PE, Huff DR (1995) Evolutionary implications of allozyme and RAPD variation in diploid populations of dioecious buffalograss Buchloe dactyloides. Mol Ecol 4:135–147
Peakall R, Ruibal M, Lindenmayer DB (2003) Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution 57:1182–1195
Peakall R, Smouse PE (2005) Program note GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 10.1111/j.1471–8286.2005.01155.x
Pech RP, Hone J (1988) A model of the dynamics and control of an outbreak of foot-and-mouth disease in feral pigs in Australia. J Appl Ecol 25:63–77
Piry S, Luikart G, Cornuet JM (1999) A computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen W (2004) Documentation for Sructure: Version 2. University of Chicago, Chicago, IL, USA
Raymond M, Rousset F (1995) GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Saunders G, Bryant H (1988) The evaluation of a feral pig eradication program during a simulated exotic disease outbreak. Aust Wildl Res 15:73–81
Simard F, Fontenille D, Lehmann T, Girod R, Brutus L, Gopaul R, Dournon C, Collins FH (1999) High amounts of genetic differentiation between populations of the Malaria vector Anopheles arabiensis from west Africa and eastern outer islands. Am J Trop Med Hygeine 60(6):1000–1009
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47:264–279
Smouse PE, Peakall R (1999) Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82:561–573
Smyth AK, James CD (2004) Characteristics of Australia’s rangelands and key design issues for monitoring biodiversity. Aust Ecol 29:3–15
Spencer PBS, Lapidge SJ, Hampton JO, Pluske JR (2005) The sociogenetic structure of a controlled feral pig population. Wildl Res 32:297–304
Spencer PB, Woolnough AP (2004) Size should matter: distribution and genetic considerations for pest animal management. Ecol Manage Restor 5:231–232
ter Braak CJF (1986) Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67:11167–1179
ter Braak CJF, Smilauer P (2002) CANOCO reference manual and canodraw for windows users guide: software for canonical community ordination (Version 4.5). Microcomputer power (Ithaca, NY, USA), p 500
ter Braak CJE, Verdonschot PEM (1995) Canonical correspondence analysis and related multivariate methods in aquatic ecology. Aquat Sci 57:255–289
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Program note: micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Vernesi C, Crestanello B, Pecchioli E, Tartari D, Caramelli D, Hauffe H, Bertorelle G (2003) The genetic impact of demographic decline and reintroduction in the wild boar (Sus scrofa): a microsatellite analysis. Mol Ecol 12:585–595
Weber JL, Wong C (1993) Mutation of short tandem repeats. Hum Mol Genet 2:1123–1128
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wright S (1943) Isolation by distance. Genetics 28:114–138
Wright S (1978) Evolution and the Genetics of Populations. Vol. IV. Variability Within and Among Natural Populations. University of Chicago Press, Chicago
Yeh F, Yang R, Boyle T (1999) PopGene 2 (Version 1.31): Microsoft windows-based freeware for population genetic analysis. University of Alberta and Centre for International Forestry Research. Available from http://www.ualberta.ca/~fyeh/info.htm
Acknowledgements
This project was funded by the NHT through the NFACP (Commonwealth Government Bureau of Rural Sciences) and a Meat and Livestock Australia Scholarship to the corresponding author. Thanks to the Invasive Animals Cooperative Research Centre for additional support. Thanks to Peter Spencer for advice on sampling, the many hunters (especially Alan Brady) for contributing samples, the QPWS for access to Welford National Park, Tony McManus and the Clyde Agricultural Co. for station access, Steve Sarre for comments on an earlier manuscript, and the Bureau of Meteorology, Geoscience Australia and Qld DNRM for geographic and environmental data. The Qld DNRM Pest Animal Ethics Committee and the NSW DPI AEC approved research for associated animal use in which samples were collected.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cowled, B.D., Aldenhoven, J., Odeh, I.O.A. et al. Feral pig population structuring in the rangelands of eastern Australia: applications for designing adaptive management units. Conserv Genet 9, 211–224 (2008). https://doi.org/10.1007/s10592-007-9331-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-007-9331-1