Abstract
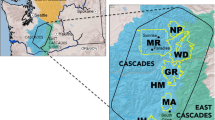
To counter losses of genetic diversity in reintroduced populations, species sometimes are reintroduced into networks of populations with the potential to exchange individuals. In reintroduced populations connected by gene flow, patterns of genetic structure initiated by the founding event may become obscured, and populations may eventually follow an isolation-by-distance model of genetic differentiation. Taking advantage of well-documented reintroduction histories of wild turkey populations in Indiana, we assessed the degree to which gene flow among reintroduced populations has obscured genetic signatures left by the founding events. Using a suite of nuclear microsatellite loci and sequence data from the mitochondrial control region, we characterized the level of genetic diversity and degree of genetic structure within and among: (1) reintroduced populations in isolated northern Indiana Fish and Wildlife Areas, (2) reintroduced populations in southern Indiana Fish and Wildlife Areas, where the distribution of populations is more continuous, and (3) source populations used for these reintroductions. We also utilized individual-based assignment tests to determine the relative contribution of source populations to the current distribution of alleles in reintroduced populations. Our results indicate that wild turkey reintroductions in Indiana have left distinct genetic signatures on populations that are detectable even after several decades. Although we found some case-specific evidence for gene flow, particularly in regions where populations are in close proximity, our data indicate on overall paucity of gene flow at a regional scale. Such post-reintroduction genetic monitoring has immediate implications for the design of optimal strategies to reintroduce wildlife for conservation and management.
Similar content being viewed by others
References
Allendorf FW (1983) Isolation, gene flow, and genetic differentiation among populations. In: Schonewald-Cox CM, Chambers SM, MacBryde B, Thomas L (eds), Genetics and conservation: a reference for managing wild animal and plant populations. Benjamin/Cummings, Menlo Park, CA, pp. 51–65
Backs SE, Eisfelder CH (1990) Criteria and guidelines for wild turkey release priorities in Indiana. Proceedings of the National Wild Turkey Symposium 6:134–143
Baker AJ, Moeed A (1987) Rapid genetic differentiation and founder effect in colonizing populations of common mynahs (Acridotheres tristis). Evolution 41:525–538
Baker AJ (1992) Genetic and morphometric divergence in ancestral European and descendent New Zealand populations of chaffinches (Fringella coelebs). Evolution 46:1784–1800
Depaulis F, Veuille M (1998). Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol. Biol. Evol. 15:1788–1790
Fitzsimmons NN, Buskirk SW, Smith MH (1997) Genetic changes in reintroduced Rocky Mountain bighorn sheep populations. J. Wildlife Manag. 61:863–872
Fuerst PA, Maruyama T (1986) Considerations on the conservation of alleles and of genic heterozygosity in small managed populations. Zoo Biol. 5:171–179
Glaubitz JC (2004) CONVERT (version 1.2): A user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol. Ecol. Notes 4:309–310
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html.
Hardy OJ, Vekemans X (2002). SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2:618–620
Holm S (1979) A simple sequentially rejective multiple test procedure. Scand. J. Statist., 6, 65–70.
Huang HB, Song YQ, Hsel M, Zahorchak R, Chiu J, Teuscher C, Smith EJ (1999) Development and characterization of genetic mapping resources for the turkey (Meleagris gallopavo). J. Hered. 90:240–242
Hurlbert SH (1971) The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577–586
Latch EK, Smith EJ, Rhodes OE (2002) Isolation and characterization of microsatellite loci in wild and domestic turkeys (Meleagris gallopavo). Mol. Ecol. Notes 2:176–178
Latch EK (2004) Population genetics of reintroduced wild turkeys: insights into hybridization, gene flow, and social structure. Dissertation, Purdue University, West Lafayette, IN
Latch EK, King JS, Harveson LA, Hobson MD, Rhodes OE (2005) Assessing hybridization in wildlife populations using molecular markers: A case study in wild turkeys. J. Wildlife Manag., in press.
Leberg PL (1991) Effects of bottlenecks on genetic divergence in populations of the wild turkey. Conserv. Biol. 5:522–530
Leberg PL, Stangel PW, Hillestad HO, Marchinton RL, Smith MH (1994) Genetic structure of reintroduced wild turkey and white-tailed deer populations. J. Wildlife Manag. 58:698–711
Leberg PL, Ellsworth DL (1999) Further evaluation of the genetic consequences of translocations on southeastern white-tailed deer populations. J. Wildlife Manag. 63:327–334
Lewis PO, Zaykin D (1999) Genetic data analysis: a computer program for the analysis of allelic data, version 1.1. Available at http://lewis.eeb.uconn.edu/lewishome/software.html.
Luikart G, Allendorf FW, Cornuet JM, Sherwin WB (1998a) Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Hered. 89:238–247
Luikart G, Sherwin WB, Steele BM, Allendorf FW (1998b) Usefulness of molecular markers for detecting population bottlenecks via monitoring genetic change. Mol. Ecol. 7:963–974
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209–220
Maruyama T, Fuerst PA (1985) Population bottlenecks and nonequilibrium models in population genetics. 2. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111:675–689
Merila J, Bjorklund M, Baker A (1996) The successful founder: genetics of introduced Caruelis chloris (greenfinch) populations in New Zealand. Heredity 77:410–422
Mock KM, Latch EK, Rhodes OE (2004) Assessing losses of genetic diversity due to translocations: long-term case histories in Merriam’s turkey (Meleagris gallopavo merriami). Conserv. Genet. 5:631–645
Nei M, Li W-H (1973) Linkage disequilibrium in subdivided populations. Genetics 75:213–219
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Nei M (1987) Molecular Evolutionary Genetics. Columbia University Press, New York
Page RDM (1996) TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358
Pearce J, Fields RL, Scribner KT (1997) Nest materials as a source of genetic data for avian behavioral studies. J. Field Ornithol., 68, 471–481
Perez T, Albornoz J, Nores C, Dominguez A (1998) Evaluation of genetic variability in introduced populations of red deer (Cervus elaphus) using DNA fingerprinting. Hereditas 129:85–89
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 12:844–855
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (1995) GENEPOP (version 1.2): Population-genetics software for exact tests and ecumenicism. J. Hered. 86:248–249
Rhodes OE, Buford DJ, Miller MS, Lutz RS (1995) Genetic structure of reintroduced Rio Grande wild turkeys in Kansas. J. Wildlife Manag. 59:771–775
Rhodes OE, Reat EP, Heffelfinger JR, DeVos JC (2001) Analysis of reintroduced pronghorn populations in Arizona using mitochondrial DNA markers. Proceedings of the Biennial Pronghorn Antelope Workshop 19:45–54
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Rowe GT, Beebee JC, Burke T (1998) Phylogeography of the natterjack toad Bufo calamita in Britain: genetic differentiation of native and translocated populations. Mol. Ecol. 7:751–760
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Scribner KT, Stuwe M (1994) Genetic relationships among alpine ibex Capra ibex populations re-established from a common ancestral source. Biol. Conserv. 69:137–143
Slatkin M (1993) Isolation by distance in equilibrium and nonequilibrium populations. Evolution 47:264–279
Sokal RR, Wartenberg DE (1983) A test of spatial auto-correlation analysis using an isolation-by-distance model. Genetics 105:219–237
Tajima F (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics 105:437–460
Weir BS, Cockerham CC (1984) Estimating F-Statistics for analysis of population structure. Evolution 38, 1358–1370
Williams RN, Rhodes OE, Serfass TL (2000) Assessment of genetic variance among source and reintroduced fisher populations. J. Mammal. 81:895–907
Williams CL, Serfass TL, Cogan R, Rhodes OE (2002) Microsatellite variation in the reintroduced Pennsylvania elk herd. Mol. Ecol. 11:1299–1310
Wright S (1978) Evolution and the genetics of populations, Vol. 4: variability within and among natural populations. University of Chicago Press, Chicago, IL
Acknowledgements
The authors are grateful for the technical assistance provided by Courtney Shattuck and Rochelle Jacques. We are indebted to Steve Backs from the Indiana Department of Natural Resources for his expertise in turkey biology and the management history of turkeys in Indiana, and to the numerous check station operators and wildlife biologists throughout our sampling area who helped us collect samples. We also would like to thank Jeff Glaubitz, Paul Leberg, Andrew DeWoody, and George Parker for helpful comments on earlier versions of the manuscript. We appreciate funding provided by Purdue University, the National Wild Turkey Federation, and the John S. Wright Endowment to the Department of Forestry and Natural Resources, Purdue University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Latch, E.K., Rhodes, O.E. The effects of gene flow and population isolation on the genetic structure of␣reintroduced wild turkey populations: Are genetic signatures of source populations retained?. Conserv Genet 6, 981–997 (2005). https://doi.org/10.1007/s10592-005-9089-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-005-9089-2