Abstract
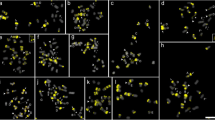
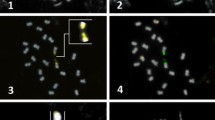
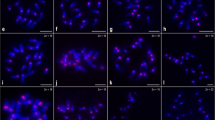
Most species of Citrus and related genera display a similar karyotype with 2n = 18 and a variable number of terminal heterochromatic blocks positively stained with chromomycin A3 (CMA+ bands). Some of these blocks are 45S rDNA sites, whereas others may correspond to the main GC-rich satellite DNA found in several Citrus species. In the present work, the distribution of the 45S rDNA and the main satellite DNA isolated from C. sinensis (CsSat) were investigated by in situ hybridization in seven species of Citrus, two species of closely related genera (Fortunella obovata and Poncirus trifoliata) and four species of the subfamily Aurantioideae, which were less related to Citrus (Atalantia monophylla, Murraya paniculata, Severinia buxifolia, and Triphasia trifolia). In Citrus, Fortunella, and Poncirus, most CMA+ bands colocalized only with CsSat sites, whereas others colocalized only with rDNA sites. However, some of these species displayed a few CMA+ bands that colocalized with sites of both probes and other CMA+ bands that did not colocalized with any of the probes. On the other hand, in the four species less related to Citrus, no CsSat signal was found on chromosomes. On Southern blot, the CsSat probe hybridized with genomic DNA from Citrus, Fortunella, and Poncirus at high stringency only, while under the less stringent conditions, it also hybridized with distantly related species. Therefore, CsSat sequences are the principal component of the heterochromatic blocks of Citrus, Poncirus, and Fortunella, whereas CsSat-like sequences seem to be widespread in the subfamily Aurantioideae. These data further suggest that the variable number of terminal CMA+ bands observed on chromosomes of Citrus and related genera are probably the consequence of amplification or reduction in the number of CsSat-like sequences distributed on chromosome termini, paralleled by mutation and homogenization events, as proposed by the library hypothesis.






Similar content being viewed by others
Abbreviations
- CMA:
-
Chromomycin A3
- DAPI:
-
4′,6-diamidino-2-phenylindole
- FISH:
-
Fluorescence in situ hybridization
- FITC:
-
Fluorescein isothiocyanate
- CsSat:
-
Citrus sinensis satellite DNA
- PCR:
-
Polymerase chain reaction
References
Beridze T, Tsirekidze N, Roytberg MA (1992) On the tertiary structure of satellite DNA. Biochimie 74:187–194
Brasileiro-Vidal AC, dos Santos-Serejo JA, Soares Filho WS, Guerra M (2007) A simple chromosomal marker can reliably distinguishes Poncirus from Citrus species. Genetica 129:273–279
Carvalho R, Soares Filho WS, Brasileiro-Vidal AC, Guerra M (2005) The relationship among lemons, limes and citron: a chromosomal comparison. Cytogenet Genome Res 109:276–282
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
Cornélio MTMN, Figueirôa ARS, Santos KGB, Soares Filho WS, Guerra M (2003) Chromosomal relationships among cultivars of Citrus reticulata Blanco, its hybrids and related species. Plant Syst Evol 240:149–161
CSHL/WUGS/PEB (2000) The complete sequence of a heterochromatic island from a higher eukaryote. The Cold Spring Harbor Laboratory, Washington University Genome Sequencing Center, and PE Biosystems Arabidopsis Sequencing Consortium. Cell 100:377–386
De Felice B, Wilson RR, Ciarmiello L, Scarano MT, Ferrante S (2006) Characterization of a novel satellite DNA sequence from flying dragon (Poncirus trifoliata). Genetica 127:45–53
Fann JY, Kovarik A, Hemleben V, Tsirekidze NI, Beridze TG (2001) Molecular and structural evolution of Citrus satellite DNA. Theor Appl Genet 103:1068–1073
Fry K, Salser W (1977) Nucleotide sequences of HS-alpha satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell 2:1069–1084
Guerra M (1993a) High amount of heterochromatin in a tropical tree species: Genipa americana L. (Rubiaceae). Cytologia 58:427–432
Guerra M (1993b) Cytogenetics of Rutaceae. V. High chromosomal variability in Citrus species revealed by CMA/DAPI staining. Heredity 71:234–241
Guerra M (2009) Chromosomal variability and the origin of Citrus species. In: Mahoney CL, Springer DA (eds) Genetic diversity. Nova Science, New York, pp 51–68
Guerra M, Santos KGB, Barros e Silva AE, Ehrendorfer F (2000) Heterochromatin banding patterns in Rutaceae–Aurantioideae—a case of parallel chromosomal evolution. Am J Bot 87:735–747
Hemleben V, Kovarik A, Torres-Ruiz RA, Volkov RA, Beridze T (2007) Plant highly repeated satellite DNA: molecular evolution, distribution and use for identification of hybrids. Syst Biodivers 5:277–289
Hou MH, Robinson H, Gao YG, Wang AH (2004) Crystal structure of the [Mg2+-(chromomycin A3)2]-d(TTGGCCAA)2 complex reveals GGCC binding specificity of the drug dimmer chelated by a metal ion. Nucleic Acids Res 32:2214–2222
Ingle J, Pearson GG, Sinclair J (1973) Species distribution and properties of nuclear satellite DNA in higher plants. Nature 242:193–197
Kang SK, Lee DH, An HY, Park JH, Yun SH, Moon YE, Bang JW, Hur Y, Hoe Koo DH (2008) Extensive chromosomal polymorphism revealed by ribosomal DNA and satellite DNA loci in 13 Citrus species. Mol Cells 26:319–322
Kazama Y, Sugiyama R, Suto Y, Uchida W, Kawano S (2006) The clustering of four subfamilies of satellite DNA at individual chromosome ends in Silene latifolia. Genome 49:520–530
Lim KY, Kovarik A, Matyasek R, Chase MW, Knapp S, McCarthy E, Clarkson JJ, Leitch AR (2006) Comparative genomics and repetitive sequence divergence in the species of diploid Nicotiana section Alatae. Plant J 48:907–919
Moraes AP, Soares Filho WS, Guerra M (2007a) Karyotype diversity and the origin of grapefruit. Chromosome Res 15:115–121
Moraes AP, Lemos RR, Brasileiro-Vidal AC, Soares Filho WS, Guerra M (2007b) Chromosomal markers distinguish hybrids and non-hybrid accessions of mandarin. Cytogenet Genome Res 119:275–181
Matsuyama T, Akihama T, Ito Y, Omura M, Fukui K (1996) Characterization of heterochromatic regions in ‘Trovita’ orange (Citrus sinensis Osbeck) chromosomes by the fluorescent staining and FISH method. Genome 39:941–945
Miranda M, Ikeda F, Endo T, Moriguchi T, Omura M (1997) Comparative analysis on the distribution of heterochromatin in Citrus, Poncirus and Fortunella chromosomes. Chromosome Res 5:86–92
Morton CM, Grant M, Blackmore S (2003) Phylogenetic relationships of the Aurantioideae inferred from chloroplast DNA sequence data. Am J Bot 90:1463–1469
Pedrosa A, Schweizer D, Guerra M (2000) Cytological heterozygosity and the hybrid origin of sweet orange [Citrus sinensis (L.) Osbeck]. Theor Appl Genet 100:361–367
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Schwarzacher T (2003) DNA, chromosomes, and in situ hybridization. Genome 46:953–962
Sharma S, Raina SN (2005) Organization and evolution of highly repeated satellite DNA sequences in plant chromosomes. Cytogenet Genome Res 109:15–26
Swingle WT, Reece PC (1967) The botany of Citrus and its wild relatives. In: Reuther W, Weber HJ, Batchelor LD (eds) The Citrus industry. History, world distribution, botany and varieties, vol I. University of California Press, Berkeley, pp 190–430
Tek AL, Song J, Macas J, Jiang J (2005) Sobo, a recently amplified satellite repeat of potato, and its implications for the origin of tandemly repeated sequences. Genetics 170:1231–1238
Wanzenböck EM, Schöfer C, Schweizer D, Bachmair A (1997) Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana. Plant J 11:1007–1016
Winterfeld G, Röser M (2007) Chromosomal localization and evolution of satellite DNAs and heterochromatin in grasses (Poaceae), especially tribe Aveneae. Plant Syst Evol 264:75–100
Yamamoto M, Abkenar AA, Matsumoto R, Kubo T, Tominaga S (2008) CMA staining analysis of chromosomes in Citrus relatives, Clymenia, Eremocitrus and Microcitrus. J Jpn Soc Hortic Sci 77:24–27
Zellinger B, Akimcheva S, Puizina J, Schirato M, Riha K (2007) Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Cell 27:163–169
Acknowledgements
This research was supported by grant of the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE). We thank Dr. Walter dos Santos Soares Filho, from EMBRAPA/CNPMF, who kindly send us seeds from several species, Andrea Pedrosa-Harand for critical comments, and Gabriela Cabral for improving the English.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Pat Heslop - Harrison.
Rights and permissions
About this article
Cite this article
Barros e Silva, A.E., Marques, A., dos Santos, K.G.B. et al. The evolution of CMA bands in Citrus and related genera. Chromosome Res 18, 503–514 (2010). https://doi.org/10.1007/s10577-010-9130-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-010-9130-2