Abstract
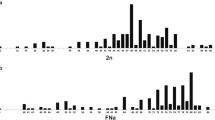
The evolutionary success of rodents of the superfamily Muroidea makes this taxon the most interesting for evolution studies, including study at the chromosomal level. Chromosome-specific painting probes from the Chinese hamster and the Syrian (golden) hamster were used to delimit homologous chromosomal segments among 15 hamster species from eight genera: Allocricetulus, Calomyscus, Cricetulus, Cricetus, Mesocricetus, Peromyscus, Phodopus and Tscherskia (Cricetidae, Muroidea, Rodentia). Based on results of chromosome painting and G-banding, comparative maps between 20 rodent species have been established. The integrated maps demonstrate a high level of karyotype conservation among species in the Cricetus group (Cricetus, Cricetulus, Allocricetulus) with Tscherskia as its sister group. Species within the genera Mesocricetus and Phodopus also show a high degree of chromosomal conservation. Our results substantiate many of the conclusions suggested by other data and strengthen the topology of the Muroidea phylogenetic tree through the inclusion of genome-wide chromosome rearrangements. The derivation of the muroids karyotypes from the putative ancestral state involved centric fusions, fissions, addition of heterochromatic arms and a great number of inversions. Our results provide further insights into the karyotype relationships of all species investigated.
Similar content being viewed by others
References
Adkins RM, Walton AH, Honeycutt RL (2002) Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol Phylogenet Evol 26: 409–420.
Aniskin VM, Benazzou T, Biltueva L, Dobigny G, Granjon L, Volobouev V (2006) Unusually extensive karyotype reorganization in four congeneric Gerbillus species (Muridae: Gerbillinae). Cytogenet Genome Res 112: 131–140.
Carleton MD, Musser GG (2005) Order Rodentia. In Wilson E, Reeder D-AM, eds., Mammal Species of the World: a Taxonomic and Geographic Reference. Baltimore: Johns Hopkins University Press, pp. 926–930, 1039–1186.
Cavagna P, Stone G, Stanyon R (2002) Black rat (Rattus rattus) genomic variability characterized by chromosome painting. Mamm Genome 13: 157–163.
Dobigny G, Ducroz JF, Robinson TJ, Volobouev V (2004) Cytogenetics and cladistics. Syst Biol 53: 470–484.
Engelbrecht A, Dobigny G, Robinson TJ (2006) Further insights into the ancestral murine karyotype: the contribution of the Otomys–Mus comparison using chromosome painting. Cytogenet Genome Res 112: 126–130.
Ferguson-Smith MA, Yang F, O’Brien PC (1998) Comparative mapping using chromosome sorting and painting. ILAR J 39: 68–76.
Froenicke L, Caldes MG, Graphodatsky A et al. (2006) Are molecular cytogenetics and bioinformatics suggesting diverging models of ancestral mammalian genomes? Genome Res 16: 306–310.
Gamperl R, Vistorin G, Rosenkranz W (1978) Comparison of chromosome banding patterns in five members of Cricetinae with comments on possible relationships. Caryologia 31: 343–353.
Graphodatsky AS (1989) Conserved and variable elements of mammalian chromosomes. In Halnan CRE, ed., Cytogenetics of Animals. Oxford: CAB International Press, pp. 95–123.
Graphodatsky AS, Sablina OV, Meyer MN et al. (2000) Comparative cytogenetics of hamsters of the genus Calomyscus. Cytogenet Cell Genet 88: 296–304.
Graphodatsky AS, Yang F, O’Brien PCM et al. (2001) Phylogenetic implications of the 38 putative ancestral chromosome segments for four canid species. Cytogenet Cell Genet 92: 243–247.
Guilly M-N, Dano L, de Chamisso P, Fouchet P, Dutrillaux B, Chevillard S (2001) Comparative karyotype using bidirectional chromosome painting: how and why? Methods Cell Sci 23: 163–170.
Guilly M-N, Fouchet P, de Chamisso P, Schmitz A, Dutrillaux B (1999) Comparative karyotype of rat and mouse using bidirectional chromosome painting. Chromosome Res 7: 213–221.
Jansa SA, Weksler M (2004) Phylogeny of muroid rodents: relationships within and among major lineages as determined by IRBP gene sequences. Mol Phylogenet Evol 31: 256–276.
Li T, O’Brien PC, Biltueva L et al. (2004) Evolution of genome organizations of squirrels (Sciuridae) revealed by cross-species chromosome painting. Chromosome Res 12: 317–335.
Li T, Wang J, Su W, Nie W, Yang F (2006a) Karyotypic evolution of the family Sciuridae: inferences from the genome organizations of ground squirrels. Cytogenet Genome Res 112: 270–276.
Li T, Wang J, Su W, Yang F (2006b) Chromosomal mechanisms underlying the karyotype evolution of the oriental voles (Muridae, Eothenomys). Cytogenet Genome Res 114: 50–55.
Matsubara K, Nishuda-Umehara C, Tsuchiya K, Nukaya D, Matsuda Y (2004) Karyotypic evolution of Apodemus (Muridae, Rodentia) inferred from comparative FISH analyses. Chromosome Res 12: 383–395.
Matsubara K, Nishida-Umehara C, Kuriowa A, Tsuchiya K, Matsuda Y (2003) Identification of chromosome rearrangements between the laboratory mouse (Mus musculus) and the Indian spiny mouse (Mus platythrix) by comparative FISH analysis. Chromosome Res 11: 57–64.
Michaux J, Reyes A, Catzeftis F (2001) Evolutionary history of the most specious mammals: molecular phylogeny of muroid rodents. Mol Biol Evol 18: 2017–2031.
Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, O’Brien SJ (2001a) Molecular phylogenetics and the origins of placental mammals. Nature 409: 614–618.
Murphy WJ, Larkin DM, Everts-van der Wind A et al. (2005) Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309: 613–617.
Murphy WJ, Stanyon, O’Brien SJ (2001b) Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biol 2: 1–8.
Neumann K, Michaux J, Lebedev V et al. (2006) Molecular phylogeny of the Cricetinae subfamily based on the mitochondrial cytochrome b and 12S rRNA genes and the nuclear vWF gene. Mol Phylogenet Evol 39: 135–148.
Radjabli SI (1975) The karyotypic differentiation of Palaearctic hamsters (Rodentia, Cricetidae). Reports of AS of USSR 225: 697–700.
Radjabli SI, Sablina OV, Graphodatsky AS (2006) Selected karyotypes. In O’Brien SJ, Nash WG, Menninger JC, eds., ATLAS of Mammalian Karyotypes. Chichester: John Wiley, pp. 188, 200–221.
Rambau RV, Robinson TJ (2003) Chromosome painting in the African four-striped mouse Rhabdomys pumilio: detection of possible murid specific contiguous segment combination. Chromosome Res 11: 91–98.
Richard F, Messaoudi C, Bonnet-Gamier A, Lombard M, Dutrillaux B (2003) Highly conserved chromosomes in an Asian squirrel (Menetes berdmorei, Rodentia: Sciuridae) as demonstrated by ZOO-FISH with human probes. Chromosome Res 11: 597–603.
Romanenko SA, Perelman PL, Serdukova NA et al. (2006) Reciprocal chromosome painting between three laboratory rodent species. Mamm Genome 17: 1183–1192.
Schmid M, Haaf T, Weis H, Schempp W (1986) Chromosomal homoeologies in hamster species of the genus Phodopus (Rodentia, Cricetinae). Cytogenet Cell Genet 43: 168–173.
Seabright M (1971) A rapid banding technique for human chromosomes. Lancet 2: 971–972.
Stanyon R, Stone G, Garcia M, Froenicke L (2003) Reciprocal chromosome painting shows that squirrels, unlike murid rodents, have a highly conserved genome organization. Genomics 82: 245–249.
Stanyon R, Yang F, Cavagna P et al. (1999) Reciprocal chromosome painting shows that genomic rearrangement between rat and mouse proceeds ten times faster than between humans and cats. Cytogenet Cell Genet 84: 150–155.
Stanyon R, Yang F, Morescalchi AM, Galleni L (2004) Chromosome painting in the long-tailed field mouse provides insights into the ancestral murid karyotype. Cytogenet Genome Res 105: 406–411.
Steppan SJ, Adkins RM, Anderson J (2004) Phylogeny and divergence-data estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst Biol 53: 533–553.
Swofford DL (1998) PAUP: Phylogenetic Analysis Using Parcimony, version 4.0. Sinauer Associates, Sunderland, MA.
Veyrunes F, Dobigny G, Yang F et al. (2006) Phylogenomics of the genus Mus (Rodentia; Muridae): extensive genome repatterning is not restricted to the house mouse. Proc R Soc B 273: 2925–2934.
Viegas-Pequignot E, Petit D, Benazzou T et al. (1986) Chromosomal evolution in rodents. Mammalia 50: 164–202.
Volobouev VT, Gallardo MH, Graphodatsky AS (2006) Rodents cytogenetics. In O’Brien J, Nash WG, Menninger JC, eds., ATLAS of Mammalian Karyotypes. Chichester: John Wiley, pp. 173–176.
Vorontzov NN, Potapova EG (1979) Taxonomy of the genus Calomyscus: status of the Calomyscus in the system of Cricetidae. Zool J 58: 1391–1397 [Rusian].
Yang F, Fu B, O’Brien PC, Robinson TJ, Ryder OA, Ferguson-Smith MA (2003) Karyotypic relationships of horses and zebras: results of cross-species chromosome painting. Cytogenet Genome Res 102: 235–243.
Yang F, O’Brien PC, Milne BS (1999) A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics 62: 189–202.
Yang F, O’Brien PCM, Ferguson-Smith MA (2000) Comparative chromosome map of the laboratory mouse and Chinese hamster defined by reciprocal chromosome painting. Chromosome Res 8: 219–227.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Romanenko, S.A., Volobouev, V.T., Perelman, P.L. et al. Karyotype evolution and phylogenetic relationships of hamsters (Cricetidae, Muroidea, Rodentia) inferred from chromosomal painting and banding comparison. Chromosome Res 15, 283–298 (2007). https://doi.org/10.1007/s10577-007-1124-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-007-1124-3