Abstract
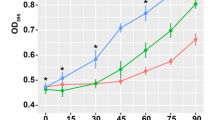
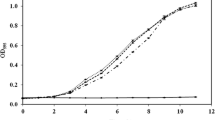
Many contaminated sites are characterized by the presence of different metals, thus increasing the complexity of toxic responses in exposed organisms. Within toxicogenomics, transcriptomics can be approached through the use of microarrays aimed at producing a genetic fingerprint for the response of model organisms to the presence of chemicals. We studied temporal changes in the early gene expression profiles of Escherichia coli cells exposed to three metal doses of a polymetallic solution over three exposure times, through the application of cDNA microarray technology. In the absence of metals, many genes belonging to a variety of cellular functions were up- and down-regulated over time. At the lowest metal dose, an activation of metal-specific transporters (Cus and ZraP proteins) and a mobilization of glutathione transporters involved in metal sequestration and trafficking was observed over time; this metal dose resulted in the generation of ROS capable of stimulating the transcription of Mn-superoxide dismutase, the assembly of Fe-S clusters and the synthesis of cysteine. At the intermediate dose, an overexpression of ROS scavengers (AhpF, KatG, and YaaA) and heat shock proteins (ClpP, HslV, DnaK, and IbpAB) was observed. Finally, at the highest dose, E. coli cells showed a repression of genes related with DNA mutation correctors (MutY glycopeptidases).




Similar content being viewed by others
References
Addis P, Shecterle LM, St. Cyr JA. Cellular protection during oxidative stress: a potential role for D-ribose and antioxidants. J Diet Suppl. 2012;9:178–82.
Agrawal SB, Agrawal M, Lee EH, Kramer GF, Pillai P. Changes in polyamine and glutathione contents of a green alga, Chlorogonium elongatum (Dang) France exposed to mercury. Environ Exp Bot. 1992;32:145–51.
Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50.
Aryee M, Gutierrez-Pabello J, Kramnik I, Maiti T, Quackenbush J. An improved empirical Bayes approach to estimating differential gene expression in microarray time-course data: BETR (Bayesian Estimation of Temporal Regulation). BMC Bioinforma. 2009;10:409.
Asad NR, Asad L, de Almeida CEB, Felzenszwalb I, Cabral-Neto JB, Leitão AC. Several pathways of hydrogen peroxide action that damage the Escherichia coli genome. Genet Mol Biol. 2004;27:291–303.
Baek YW, An YJ. Microbial toxicity of metal oxide nanoparticles (CuO, NiO, ZnO, and Sb2O3) to Escherichia coli, Bacillus subtilis, and Streptococcus aureus. Sci Total Environ. 2011;409:1603–8.
Bagai I, Liu W, Rensing C, Blackburn NJ, McEvoy MM. Substrate-linked conformational change in the periplasmic component of a Cu(I)/Ag(I) efflux system. J Biol Chem. 2007;282:35695–702.
Barras F, Fontecave M. Cobalt stress in Escherichia coli and Salmonella enterica: Molecular bases for toxicity and resistance. Metallomics. 2011;3:1130–4.
Bolstad BM, Irizarry RA, Åstrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93.
Booth IR, Blount P. The MscS and MscL families of mechanosensitive channels act as microbial emergency release valves. J Bacteriol. 2012;194:4802–9.
Bruins MR, Kapil S, Oehme FW. Microbial resistance to metals in the environment. Ecotoxicol Environ Saf. 2000;45:198–207.
Bruno-Barcena JM, Azcarate-Peril MA, Hassan HM. Role of antioxidant enzymes in bacterial resistance to organic acids. Appl Environ Microbiol. 2010;76:2747–53.
Chassagnole C, Quentin E, Fell DA, de Atauri P, Mazat JP. Dynamic simulation of pollutant effects on the threonine pathway in Escherichia coli. C R Biol. 2003;326:501–8.
Cizewski Culotta V, Joh H-D, Lin S-J, Hudak Slekar K, Strain J. A physiological role for Saccharomyces cerevisiae copper/zinc superoxide dismutase in copper buffering. J Biol Chem. 1995;270:29991–7.
Cui J, Kaandorp JA, Lloyd CM. Simulating in vitro transcriptional response of zinc homeostasis system in Escherichia coli. BMC Syst Biol. 2008;2:1752–0509.
Decker K, Gerhardt F, Boos W. The role of the trehalose system in regulating the maltose regulon of Escherichia coli. Mol Microbiol. 1999;32:777–88.
Eggen RIL, Behra R, Burkhardt-Holm P, Escher BI, Schweigert N. Peer reviewed: Challenges in ecotoxicology. Environ Sci Technol. 2004;38:58A–64.
Ercal N, Gurer-Orhan H, Aykin-Burns N. Toxic metals and oxidative stress part I: Mechanisms involved in metal-induced oxidative damage. Curr Top Med Chem. 2001;1:529–39.
Fantino JR, Py B, Fontecave M, Barras F. A genetic analysis of the response of Escherichia coli to cobalt stress. Environ Microbiol. 2010;12:2846–57.
Fluman N, Bibi E. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim Biophys Acta. 2009;5:738–47.
Gama-Castro S, Salgado H, Peralta-Gil M, Santos-Zavaleta A, Muniz-Rascado L, Solano-Lira H, et al. RegulonDB version 7.0: Transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units). Nucleic Acids Res. 2011;39:D98–105.
Geslin C, Llanos J, Prieur D, Jeanthon C. The manganese and iron superoxide dismutases protect Escherichia coli from heavy metal toxicity. Res Microbiol. 2001;152:901–5.
Giller KE, Witter E, McGrath SP. Toxicity of heavy metals to microorganisms and microbial processes in agricultural soils: a review. Soil Biol Biochem. 1998;30:1389–414.
Goetz AK, Singh BP, Battalora M, Breier JM, Bailey JP, Chukwudebe AC, et al. Current and future use of genomics data in toxicology: Opportunities and challenges for regulatory applications. Regul Toxicol Pharmacol. 2011;61:141–53.
Graham AI, Hunt S, Stokes SL, Bramall N, Bunch J, Cox AG, et al. Severe zinc depletion of Escherichia coli. J Biol Chem. 2009;284:18377–89.
Hantke K. Members of the Fur protein family regulate iron and zinc transport in E. coli and characteristics of the Fur-regulated FhuF protein. J Mol Microbiol Biotechnol. 2002;4:217–22.
Helbig K, Grosse C, Nies DH. Cadmium toxicity in glutathione mutants of Escherichia coli. J Bacteriol. 2008;190:5439–54.
Hu P, Brodie EL, Suzuki Y, McAdams HH, Andersen GL. Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J Bacteriol. 2005;187:8437–49.
Ivask A, Bondarenko O, Jepihhina N, Kahru A. Profiling of the reactive oxygen species-related ecotoxicity of CuO, ZnO, TiO2, silver and fullerene nanoparticles using a set of recombinant luminescent Escherichia coli strains: Differentiating the impact of particles and solubilised metals. Anal Bioanal Chem. 2010;398:701–16.
Kalantari N, Ghaffari S. Evaluation of toxicity of heavy metals for Escherichia coli growth. Iran J Environ Health. 2008;5:173–8.
Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30.
Karp PD, Riley M, Saier M, Paulsen IT, Collado-Vides J, Paley SM, et al. The EcoCyc Database. Nucleic Acids Res. 2002;30:56–8.
Kershaw CJ, Brown NL, Constantinidou C, Patel MD, Hobman JL. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology. 2005;151:1187–98.
Kimura T, Nishioka H. Intracellular generation of superoxide by copper sulphate in Escherichia coli. Mutat Res. 1997;389:237–42.
Kitagawa M, Matsumura Y, Tsuchido T. Small heat shock proteins, IbpA and IbpB, are involved in resistances to heat and superoxide stresses in Escherichia coli. FEMS Microbiol Lett. 2000;184:165–71.
Krämer U, Talke IN, Hanikenne M. Transition metal transport. FEBS Lett. 2007;581:2263–72.
Kusano T, Berberich T, Tateda C, Takahashi Y. Polyamines: Essential factors for growth and survival. Planta. 2008;228:367–81.
Lee LJ, Barrett JA, Poole RK. Genome-wide transcriptional response of chemostat-cultured Escherichia coli to zinc. J Bacteriol. 2005;187:1124–34.
Lee C, Kim J, Shin SG, Hwang S. Absolute and relative qPCR quantification of plasmid copy number in Escherichia coli. J Biotechnol. 2006;123:273–80.
Lee C, Lee S, Shin SG, Hwang S. Real-time PCR determination of rRNA gene copy number: absolute and relative quantification assays with Escherichia coli. Appl Microbiol Biotechnol. 2008;78:371–6.
Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell. 2004;16:596–615.
Liu Y, Bauer SC, Imlay JA. The YaaA protein of the Escherichia coli OxyR Regulon lessens hydrogen peroxide toxicity by diminishing the amount of intracellular unincorporated iron. J Bacteriol. 2011;193:2186–96.
Long AD, Mangalam HJ, Chan BYP, Tolleri L, Hatfield GW, Baldi P. Improved statistical inference from DNA microarray data using analysis of variance and a bayesian statistical framework. J Biol Chem. 2001;276:19937–44.
Mellies J, Thomas K, Turvey M, Evans N, Crane J, Boedeker E, et al. Zinc-induced envelope stress diminishes type III secretion in enteropathogenic Escherichia coli. BMC Microbiol. 2012;12:123.
Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46.
Moore CM, Gaballa A, Hui M, Ye RW, Helmann JD. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol Microbiol. 2005;57:27–40.
Morey JS, Ryan JC, Van Dolah FM. Microarray validation: Factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 2006;8:175–93.
Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, et al. The transcriptional landscape of the yeast whole genome defined by RNA sequencing. Science. 2008;320:1344–9.
Nagalakshmi U, Waern K, Snyder M. RNA‐Seq: a method for comprehensive transcriptome analysis. Curr Protocols Mol Biol. 2010;Chapter 4:Unit 4.11.1-13. doi: 10.1002/0471142727.mb0411s89.
Nandakumar R, Espirito SC, Madayiputhiya N, Grass G. Quantitative proteomic profiling of the Escherichia coli response to metallic copper surfaces. Biometals. 2011;24:429–44.
Neumann M, Leimkuhler S. Heavy metal ions inhibit molybdoenzyme activity by binding to the dithiolene moiety of molybdopterin in Escherichia coli. FEBS J. 2008;275:5678–89.
Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev. 2003;27:313–39.
Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: Regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60.
Panek HR, O'Brian MR. KatG is the primary detoxifier of hydrogen peroxide produced by aerobic metabolism in Bradyrhizobium japonicum. J Bacteriol. 2004;186:7874–80.
Park S, Ely RL. Candidate stress genes of Nitrosomonas europaea for monitoring inhibition of nitrification by heavy metals. Appl Environ Microbiol. 2008;74:5475–82.
Peng L, Lifang R, Hongyu X, Xi L, Chaocan Z. Study on the toxic effect of lead(II) ion on Escherichia coli. Biol Trace Elem Res. 2007;115:195–202.
Pfaffl MW. Relative quantification. Real Time qPCR. Taylor & Francis Group; 2006, pp. 63–82.
Pope MA, Porello SL, David SS. Escherichia coli apurinic-apyrimidinic endonucleases enhance the turnover of the adenine glycosylase MutY with G:A substrates. J Biol Chem. 2002;277:22605–15.
Py B, Barras F. Building Fe-S proteins: Bacterial strategies. Nat Rev Microbiol. 2010;8:436–46.
Ranquet C, Ollagnier-de-Choudens S, Loiseau L, Barras F, Fontecave M. Cobalt stress in Escherichia coli. The effect on the iron-sulfur proteins. J Biol Chem. 2007;282:30442–51.
Rispoli F, Angelov A, Badia D, Kumar A, Seal S, Shah V. Understanding the toxicity of aggregated zero valent copper nanoparticles against Escherichia coli. J Hazard Mater. 2010;180:212–6.
Robbens J, van der Ven K, Maras M, Blust R, De Coen W. Ecotoxicological risk assessment using DNA chips and cellular reporters. Trends Biotechnol. 2007;25:460–6.
Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86.
Sawers G. A novel mechanism controls anaerobic and catabolite regulation of the Escherichia coli tdc operon. Mol Microbiol. 2001;39:1285–98.
Schmidt R, Zahn R, Bukau B, Mogk A. ClpS is the recognition component for Escherichia coli substrates of the N-end rule degradation pathway. Mol Microbiol. 2009;72:506–17.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–8.
Seaver LC, Imlay JA. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J Bacteriol. 2001;183:7182–9.
Sharma SS, Dietz K-J. The significance of amino acids and amino acid-derived molecules in plant responses and adaptation to heavy metal stress. J Exp Bot. 2006;57:711–26.
Sharma SK, Goloubinoff P, Christen P. Heavy metal ions are potent inhibitors of protein folding. Biochem Biophys Res Commun. 2008;372:341–5.
Sipos K, Lange H, Fekete Z, Ullmann P, Lill R, Kispal G. Maturation of cytosolic iron-sulfur proteins requires glutathione. J Biol Chem. 2002;277:26944–9.
Soukas A, Cohen P, Socci ND, Friedman JM. Leptin-specific patterns of gene expression in white adipose tissue. Gene Dev. 2000;14:963–80.
Teitzel GM, Geddie A, De Long SK, Kirisits MJ, Whiteley M, Parsek MR. Survival and growth in the presence of elevated copper: Transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J Bacteriol. 2006;188:7242–56.
Tkachenko A, Nesterova L, Pshenichnov M. The role of the natural polyamine putrescine in defense against oxidative stress in Escherichia coli. Arch Microbiol. 2001;176:155–7.
Valls M, Lorenzo V. Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol Rev. 2002;26:327–38.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paene A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.0031–11.
Veinger L, Diamant S, Buchner J, Goloubinoff P. The small heat-shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J Biol Chem. 1998;273:11032–7.
Wang A, Crowley DE. Global gene expression responses to cadmium toxicity in Escherichia coli. J Bacteriol. 2005;187:3259–66.
Wang S, Deng K, Zaremba S, Deng X, Lin C, Wang Q, et al. Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl Environ Microbiol. 2009;75:6110–23.
Waters MD, Fostel JM. Toxicogenomics and systems toxicology: Aims and prospects. Nat Rev Genet. 2004;5:936–48.
Worden CR, Kovac WK, Dorn LA, Sandrin TR. Environmental pH affects transcriptional responses to cadmium toxicity in Escherichia coli K-12 (MG1655). FEMS Microbiol Lett. 2009;293:58–64.
Wortham B, Oliveira M, Patel C. Polyamines in bacteria: Pleiotropic effects yet specific mechanisms. In: Perry R, Fetherston J, editors. The Genus Yersinia. New York: Springer; 2007. p. 106–15.
Xu FF, Imlay JA. Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl Environ Microbiol. 2012;78:3614–21.
Yamamoto K, Ishihama A. Transcriptional response of Escherichia coli to external copper. Mol Microbiol. 2005a;56:215–27.
Yamamoto K, Ishihama A. Transcriptional response of Escherichia coli to external zinc. J Bacteriol. 2005b;187:6333–40.
Yeung KY, Haynor DR, Ruzzo WL. Validating clustering for gene expression data. Bioinformatics. 2001;17:309–18.
Zhou J, Rudd KE. EcoGene 3.0. Nucleic Acids Res. 2013;41:D613–24.
Zimmermann M, Udagedara SR, Sze CM, Ryan TM, Howlett GJ, Xiao Z, et al. PcoE–a metal sponge expressed to the periplasm of copper resistance Escherichia coli. Implication of its function role in copper resistance. J Inorg Biochem. 2012;115:186–97.
Acknowledgments
This work has been financially supported by the following projects: 7/12/TK/2009/3 LURCHIP (Biscay County Council), BERRILUR3-Etortek (Basque Government), and MINECO AGL2012-39715-CO3-01/02. M.T. Gómez-Sagasti is the recipient of a predoctoral fellowship from the Department of Education, Universities and Research, Basque Government. Technical support by Javier Etxebarria and Amaia García from GAIKER is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gómez-Sagasti, M.T., Becerril, J.M., Martín, I. et al. cDNA microarray assessment of early gene expression profiles in Escherichia coli cells exposed to a mixture of heavy metals. Cell Biol Toxicol 30, 207–232 (2014). https://doi.org/10.1007/s10565-014-9281-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10565-014-9281-6