Abstract
Purpose
Neoadjuvant chemotherapy (NAC) is used to treat patients with high-risk breast cancer. The tumor response to NAC can be classified as either a pathological partial response (pPR) or pathological complete response (pCR), defined as complete eradication of invasive tumor cells, with a pCR conferring a significantly lower risk of recurrence. Predicting the response to NAC, however, remains a significant clinical challenge. The objective of this study was to determine if analysis of nuclear features on core biopsies using artificial intelligence (AI) can predict response to NAC.
Methods
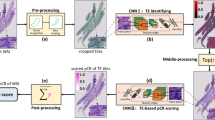
Fifty-eight HER2-positive or triple-negative breast cancer patients were included in this study (pCR n = 37, pPR n = 21). Multiple deep convolutional neural networks were developed to automate tumor detection and nuclear segmentation. Nuclear count, area, and circularity, as well as image-based first- and second-order features including mean pixel intensity and correlation of the gray-level co-occurrence matrix (GLCM-COR) were determined.
Results
In univariate analysis, the pCR group had fewer multifocal/multicentric tumors, higher nuclear intensity, and lower GLCM-COR compared to the pPR group. In multivariate binary logistic regression, tumor multifocality/multicentricity (OR = 0.14, p = 0.012), nuclear intensity (OR = 1.23, p = 0.018), and GLCM-COR (OR = 0.96, p = 0.043) were each independently associated with likelihood of achieving a pCR, and the model was able to successful classify 79% of cases (62% for pPR and 89% for pCR).
Conclusion
Analysis of tumor nuclear features using digital pathology/AI can significantly improve models to predict pathological response to NAC.




Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Code generated during the current study is available from the corresponding author on reasonable request.
References
Untch M, Konecny GE, Paepke S et al (2014) Current and future role of neoadjuvant therapy for breast cancer. Breast 23:526–537
Murphy BL, Day CN, Hoskin TL et al (2018) Neoadjuvant chemotherapy use in breast cancer is greatest in excellent responders: triple-negative and HER2+ subtypes. Ann Surg Oncol 25:2241–2248
von Minckwitz G, Untch M, Blohmer JU et al (2012) Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol 30:1796–1804
Cortazar P, Zhang L, Untch M et al (2014) Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 384:164–172
Spring LM, Fell G, Arfe A et al (2020) Pathologic complete response after neoadjuvant chemotherapy and impact on breast cancer recurrence and survival: a comprehensive meta-analysis. Clin Cancer Res 26:2838–2848
Kim KI, Lee KH, Kim TR et al (2014) Ki-67 as a predictor of response to neoadjuvant chemotherapy in breast cancer patients. J Breast Cancer 17:40–46
Tan QX, Qin QH, Yang WP et al (2014) Prognostic value of Ki67 expression in HR-negative breast cancer before and after neoadjuvant chemotherapy. Int J Clin Exp Pathol 7:6862–6870
Erbes T, Stickeler E, Rücker G et al (2016) BMI and pathologic complete response to neoadjuvant chemotherapy in breast cancer: a study and meta-analysis. Clin Breast Cancer 16:e119–e132
Matsubara N, Mukai H, Fujii S et al (2013) Different prognostic significance of Ki-67 change between pre- and post-neoadjuvant chemotherapy in various subtypes of breast cancer. Breast Cancer Res Treat 137:203–212
Jung YY, Hyun CL, Jin MS et al (2016) Histomorphological factors predicting the response to neoadjuvant chemotherapy in triple-negative breast cancer. J Breast Cancer 19:261–267
Ono M, Tsuda H, Shimizu C et al (2012) Tumor-infiltrating lymphocytes are correlated with response to neoadjuvant chemotherapy in triple-negative breast cancer. Breast Cancer Res Treat 132:793–805
Asano Y, Kashiwagi S, Goto W et al (2018) Prediction of treatment response to neoadjuvant chemotherapy in breast cancer by subtype using tumor-infiltrating lymphocytes. Anticancer Res 38:2311–2321
Kraus JA, Beriwal S, Dabbs DJ et al (2012) Predictors of pathologic complete response after standard neoadjuvant chemotherapy in triple-negative breast carcinoma. Appl Immunohistochem Mol Morphol 20:334–339
Li X, Kanbour-Shakir A, Dabbs DJ et al (2013) Morphologic features do not influence response to trastuzumab-containing neoadjuvant chemotherapy in HER2-positive breast cancer. Appl Immunohistochem Mol Morphol 21:420–425
Tran WT, Jerzak K, Lu FI et al (2019) Personalized breast cancer treatments using artificial intelligence in radiomics and pathomics. J Med Imaging Radiat Sci 50:S32–S41
Ibrahim A, Gamble P, Jaroensri R et al (2020) Artificial intelligence in digital breast pathology: techniques and applications. Breast 49:267–273
Bera K, Schalper KA, Rimm DL et al (2019) Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat Rev Clin Oncol 16:703–715
Landmann A, Farrugia DJ, Zhu L et al (2018) Low estrogen receptor (ER)-positive breast cancer and neoadjuvant systemic chemotherapy: is response similar to typical ER-positive or ER-negative disease? Am J Clin Pathol 150:34–42
Prabhu JS, Korlimarla A, Desai K et al (2014) A majority of low (1-10%) ER positive breast cancers behave like hormone receptor negative tumors. J Cancer 5:156–165
Symmans WF, Peintinger F, Hatzis C et al (2007) Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol 25:4414–4422
Vahadane A, Peng T, Sethi A et al (2016) Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans Med Imaging 35:1962–1971
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. In: Third international conference on learning representations, ICLR 2015—conference track proceedings, pp 1–14
Deng J, Dong W, Socher R et al (2009) ImageNet: a large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition, Miami, FL, pp 248–255
Nair V, Hinton EG (2010) Rectified linear units improve restricted Boltzmann machines. In: ICML’10: Proceedings of the 27th international conference on international conference on machine learning. https://dl.acm.org/doi/10.5555/3104322.3104425. Accessed 12 Oct 2020
Lin M, Chen Q, Yan S (2014) Network in network. In: Second international conference on learning representations, ICLR 2014—conference track proceedings, pp 1–10
Goodfellow I, Bengio Y, Courville A (2016) Deep learning. MIT Press, Cambridge
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. Comput Vis Pattern Recogn 9351:234–241
Dong H, Yang G, Liu F et al (2017) Automatic brain tumor detection and segmentation using U-net based fully convolutional networks. Commun Comput Inf Sci 723:506–517
He K, Zhang X, Ren S et al (2016) Deep residual learning for image recognition. In: Computer Vision and Pattern Recognition, pp 770–778
Kumar N, Verma R, Anand D et al (2020) A multi-organ nucleus segmentation challenge. IEEE Trans Med Imaging 39:1380–1391
Kumar N, Verma R, Sharma S et al (2017) A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans Med Imaging 36:1550–1560
Kumar N, Verma R, Sharma S et al (2018) Multi-organ nucleus segmentation challenge. In: International conference on medical image computing and computer-assisted intervention. https://monuseg.grand-challenge.org/Data. Accessed 12 Oct 2020
Bloice MD, Roth PM, Holzinger A (2019) Biomedical image augmentation using augmentor. Bioinformatics 35:4522–4524
Gutman DA, Khalilia M, Lee S et al (2017) The digital slide archive: a software platform for management, integration, and analysis of histology for cancer research. Cancer Res 77:e75–e78
Haralick RM, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC-3:610–621
Ataseven B, Lederer B, Blohmer JU et al (2015) Impact of multifocal or multicentric disease on surgery and locoregional, distant and overall survival of 6,134 breast cancer patients treated with neoadjuvant chemotherapy. Ann Surg Oncol 22:1118–1127
Romo-Bucheli D, Janowczyk A, Gilmore H et al (2016) Automated tubule nuclei quantification and correlation with oncotype DX risk categories in ER+ breast cancer whole slide images. Sci Rep 6:32706
Cireşan DC, Giusti A, Gambardella LM et al (2013) Mitosis detection in breast cancer histology images with deep neural networks. Med Image Comput Comput Assist Interv 16:411–418
Wang H, Cruz-Roa A, Basavanhally A et al (2014) Mitosis detection in breast cancer pathology images by combining handcrafted and convolutional neural network features. J Med Imaging (Bellingham) 1:034003
Veta M, Kornegoor R, Huisman A et al (2012) Prognostic value of automatically extracted nuclear morphometric features in whole slide images of male breast cancer. Mod Pathol 25:1559–1565
Whitney J, Corredor G, Janowczyk A et al (2018) Quantitative nuclear histomorphometry predicts oncotype DX risk categories for early stage ER+ breast cancer. BMC Cancer 18:610
Adur J, Carvalho HF, Cesar CL et al (2014) Nonlinear optical microscopy signal processing strategies in cancer. Cancer Inform 13:67–76
Lu C, Romo-Bucheli D, Wang X et al (2018) Nuclear shape and orientation features from H&E images predict survival in early-stage estrogen receptor-positive breast cancers. Lab Investig 98:1438–1448
Chen JM, Qu AP, Wang LW et al (2015) New breast cancer prognostic factors identified by computer-aided image analysis of HE stained histopathology images. Sci Rep 5:10690
Vujasinovic T, Pribic J, Kanjer K et al (2015) Gray-level co-occurrence matrix texture analysis of breast tumor images in prognosis of distant metastatis risk. Microsc Microanal 21:646–654
Acknowledgments
The authors wish to thank the Breast Site Group at Sunnybrook Health Sciences Centre for their continued support and intellectual discussions.
Funding
Dr. Lu, Dr. Sadeghi-Naini, and Dr. Tran received funding from the tri-council (Government of Canada) New Frontiers in Research Fund. Dr. Tran lab is funded in part, by the Terry Fox Research Institute and the Women’s Health Golf Classic Foundation Fund. Dr. Sadeghi-Naini holds a York Research Chair in Quantitative Imaging and Smart Biomarkers, and received funding from the Natural Sciences and Engineering Research Council of Canada and the Terry Fox Research Institute.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by DWD, AL, and ST. The first draft of the manuscript was written by DWD and AL and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Ethical approval
This study was approved by the institutional research ethics board.
Informed consent
Waived as per the institutional research ethics board.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dodington, D.W., Lagree, A., Tabbarah, S. et al. Analysis of tumor nuclear features using artificial intelligence to predict response to neoadjuvant chemotherapy in high-risk breast cancer patients. Breast Cancer Res Treat 186, 379–389 (2021). https://doi.org/10.1007/s10549-020-06093-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-020-06093-4