Abstract
Purpose
Multiple aspects of the tumor microenvironment (TME) impact breast cancer, yet the genetic modifiers of the TME are largely unknown, including those that modify tumor vascular formation and function.
Methods
To discover host TME modifiers, we developed a system called the Consomic/Congenic Xenograft Model (CXM). In CXM, human breast cancer cells are orthotopically implanted into genetically engineered consomic xenograft host strains that are derived from two parental strains with different susceptibilities to breast cancer. Because the genetic backgrounds of the xenograft host strains differ, whereas the inoculated tumor cells are the same, any phenotypic variation is due to TME-specific modifier(s) on the substituted chromosome (consomic) or subchromosomal region (congenic). Here, we assessed TME modifiers of growth, angiogenesis, and vascular function of tumors implanted in the SSIL2Rγ and SS.BN3IL2Rγ CXM strains.
Results
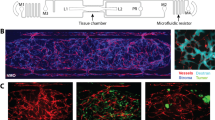
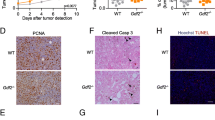
Breast cancer xenografts implanted in SS.BN3IL2Rγ (consomic) had significant tumor growth inhibition compared with SSIL2Rγ (parental control), despite a paradoxical increase in the density of blood vessels in the SS.BN3IL2Rγ tumors. We hypothesized that decreased growth of SS.BN3IL2Rγ tumors might be due to nonproductive angiogenesis. To test this possibility, SSIL2Rγ and SS.BN3IL2Rγ tumor vascular function was examined by dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI), micro-computed tomography (micro-CT), and ex vivo analysis of primary blood endothelial cells, all of which revealed altered vascular function in SS.BN3IL2Rγ tumors compared with SSIL2Rγ. Gene expression analysis also showed a dysregulated vascular signaling network in SS.BN3IL2Rγ tumors, among which DLL4 was differentially expressed and co-localized to a host TME modifier locus (Chr3: 95–131 Mb) that was identified by congenic mapping.
Conclusions
Collectively, these data suggest that host genetic modifier(s) on RNO3 induce nonproductive angiogenesis that inhibits tumor growth through the DLL4 pathway.







Similar content being viewed by others
Abbreviations
- TME:
-
Tumor microenvironment
- CXM:
-
Consomic Xenograft Model
- DCE-MRI:
-
Dynamic contrast-enhanced magnetic resonance imaging
- SSRS:
-
Species-specific RNAseq
- PBST:
-
Phosphate-buffered saline plus Tween-20
- RNO3:
-
Rat chromosome 3
- TNBC:
-
Triple-negative breast cancer
- micro-CT:
-
Micro-computed tomography
- EC:
-
Endothelial cell
- DMEM:
-
Dulbecco’s modifier Eagle’s medium
- MFP:
-
Mammary fat pad
- RARE:
-
Rapid acquisition rapid echo
- IAUC:
-
Initial area under the curve
- ROI:
-
Region of interest
- FDR:
-
False discovery rate
References
Quail DF, Joyce JA (2013) Microenvironmental regulation of tumor progression and metastasis. Nat Med 19(11):1423–1437
Polyak K, Kalluri R (2010) The role of the microenvironment in mammary gland development and cancer. Cold Spring Harbor Perspect Biol 2(11):a003244
Cook LM, Hurst DR, Welch DR (2011) Metastasis suppressors and the tumor microenvironment. Semin Cancer Biol 21(2):113–122
McAllister SS, Weinberg RA (2010) Tumor-host interactions: a far-reaching relationship. J Clin Oncol 28(26):4022–4028
Flister MJ, Endres BT, Rudemiller N, Sarkis AB, Santarriaga S, Lemke A, Roy I, Geurts AM, Moreno C, Ran S et al (2014) CXM—a new tool for mapping breast cancer risk in the tumor microenvironment. Cancer Res 74(22):6419–6429
Han M, Kim TH, Kang DK, Kim KS, Yim H (2012) Prognostic role of MRI enhancement features in patients with breast cancer: value of adjacent vessel sign and increased ipsilateral whole-breast vascularity. AJR 199(4):921–928
Craciunescu OI, Blackwell KL, Jones EL, Macfall JR, Yu D, Vujaskovic Z, Wong TZ, Liotcheva V, Rosen EL, Prosnitz LR et al (2009) DCE-MRI parameters have potential to predict response of locally advanced breast cancer patients to neoadjuvant chemotherapy and hyperthermia: a pilot study. Int J Hyperth 25(6):405–415
Ah-See ML, Makris A, Taylor NJ, Harrison M, Richman PI, Burcombe RJ, Stirling JJ, d’Arcy JA, Collins DJ, Pittam MR et al (2008) Early changes in functional dynamic magnetic resonance imaging predict for pathologic response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res 14(20):6580–6589
Jia WR, Tang L, Wang DB, Chai WM, Fei XC, He JR, Chen M, Wang WP (2016) Three-dimensional contrast-enhanced ultrasound in response assessment for breast cancer: a comparison with dynamic contrast-enhanced magnetic resonance imaging and pathology. Sci Rep 6:33832
Sun YS, He YJ, Li J, Li YL, Li XT, Lu AP, Fan ZQ, Cao K, Ouyang T (2016) Predictive value of DCE-MRI for early evaluation of pathological complete response to neoadjuvant chemotherapy in resectable primary breast cancer: a single-center prospective study. Breast 30:80–86
Heldahl MG, Bathen TF, Rydland J, Kvistad KA, Lundgren S, Gribbestad IS, Goa PE (2010) Prognostic value of pretreatment dynamic contrast-enhanced MR imaging in breast cancer patients receiving neoadjuvant chemotherapy: overall survival predicted from combined time course and volume analysis. Acta Radiol 51(6):604–612
Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, Lin HC, Yancopoulos GD, Thurston G (2006) Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature 444(7122):1032–1037
Flister MJ, Hoffman MJ, Reddy P, Jacob HJ, Moreno C (2013) Congenic mapping and sequence analysis of the Renin locus. Hypertension 61(4):850–856
Hoffman MJ, Flister MJ, Nunez L, Xiao B, Greene AS, Jacob HJ, Moreno C (2013) Female-specific hypertension Loci on rat chromosome 13. Hypertension 62(3):557–563
Volk LD, Flister MJ, Bivens CM, Stutzman A, Desai N, Trieu V, Ran S (2008) Nab-paclitaxel efficacy in the orthotopic model of human breast cancer is significantly enhanced by concurrent anti-vascular endothelial growth factor A therapy. Neoplasia 10(6):613–623
Volk-Draper LD, Rajput S, Hall KL, Wilber A, Ran S (2012) Novel model for basaloid triple-negative breast cancer: behavior in vivo and response to therapy. Neoplasia 14(10):926–942
Walker EJ, Shen F, Young WL, Su H (2011) Cerebrovascular casting of the adult mouse for 3D imaging and morphological analysis. J Vis Exp 57:e2958–e2958
Prisco AR, Bukowy JD, Hoffmann BR, Karcher JR, Exner EC, Greene AS (2014) Automated quantification reveals hyperglycemia inhibits endothelial angiogenic function. PLoS ONE 9(4):e94599
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359
Roberts A, Pachter L (2013) Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods 10(1):71–73
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11(10):R106
Flister MJ, Volk LD, Ran S (2011) Characterization of Prox1 and VEGFR-3 expression and lymphatic phenotype in normal organs of mice lacking p50 subunit of NF-kappaB. Microcirculation 18(2):85–101
Hanahan D, Folkman J (1996) Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell 86(3):353–364
Scehnet JS, Jiang W, Kumar SR, Krasnoperov V, Trindade A, Benedito R, Djokovic D, Borges C, Ley EJ, Duarte A et al (2007) Inhibition of Dll4-mediated signaling induces proliferation of immature vessels and results in poor tissue perfusion. Blood 109(11):4753–4760
Hellstrom M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N et al (2007) Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature 445(7129):776–780
Xu Z, Wang Z, Jia X, Wang L, Chen Z, Wang S, Wang M, Zhang J, Wu M (2016) MMGZ01, an anti-DLL4 monoclonal antibody, promotes nonfunctional vessels and inhibits breast tumor growth. Cancer Lett 372(1):118–127
Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I et al (2006) Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature 444(7122):1083–1087
Hoey T, Yen WC, Axelrod F, Basi J, Donigian L, Dylla S, Fitch-Bruhns M, Lazetic S, Park IK, Sato A et al (2009) DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell 5(2):168–177
Suchting S, Freitas C, le Noble F, Benedito R, Breant C, Duarte A, Eichmann A (2007) The notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc Natl Acad Sci USA 104(9):3225–3230
Siekmann AF, Lawson ND (2007) Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature 445(7129):781–784
Lobov IB, Renard RA, Papadopoulos N, Gale NW, Thurston G, Yancopoulos GD, Wiegand SJ (2007) Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc Natl Acad Sci USA 104(9):3219–3224
Bergers G, Benjamin LE (2003) Tumorigenesis and the angiogenic switch. Nat Rev Cancer 3(6):401–410
Nico B, Benagiano V, Mangieri D, Maruotti N, Vacca A, Ribatti D (2008) Evaluation of microvascular density in tumors: pro and contra. Histol Histopathol 23(5):601–607
Weidner N, Semple JP, Welch WR, Folkman J (1991) Tumor angiogenesis and metastasis–correlation in invasive breast carcinoma. N Engl J Med 324(1):1–8
Acknowledgements
We thank M. Tschannen, R. Schilling, E. Schneider, A. Zappa, and Y. Liu for excellent technical support and the Center for Imaging Research in the Medical College of Wisconsin Department of Radiology, and the Biomedical Imaging Shared Resource supported by the MCW Cancer Center.
Funding
This work was supported by a seed grant from the Wisconsin Breast Cancer Showhouse and the MCW Cancer Center, the Mary Kay Foundation (Grant No. 024.16), and the NCI (R01CA193343) to M.J.F. Support was also received from the National Center for Research Resources, the National Center for Advancing Translational Sciences, and the Office of the Director of the NIH via the Clinical & Translational Science Institute (#8KL2TR000056), the Wisconsin Breast Cancer Showhouse and the MCW Cancer Center, the Rosenberg Translational Research Award, and an institutional research Grant (#86-004-26) from the American Cancer Society to C.B.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interests to declare.
Ethical approval
All applicable international, national, and institutional guidelines for the care and use of animals were followed. The Institutional Animal Care and Use Committee (IACUC) of the Medical College of Wisconsin approved all animal studies. All procedures involving animals were conducted in accordance with the National Institutes of Health guidelines concerning the use and care of experimental animals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Flister, M.J., Tsaih, SW., Stoddard, A. et al. Host genetic modifiers of nonproductive angiogenesis inhibit breast cancer. Breast Cancer Res Treat 165, 53–64 (2017). https://doi.org/10.1007/s10549-017-4311-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-017-4311-8