Abstract
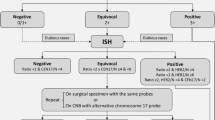
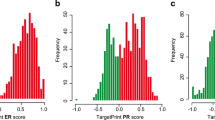
Breast cancer in African-American females (AAF) has a less favorable outcome than that in Caucasians. More information is needed regarding its biology. We evaluated gene expression in tumors from AAF presenting with early stage or locally advanced breast cancer using MammaPrint®, BluePrint ® (molecular subtype) and TargetPrint ® [estrogen receptor (ER), progesterone receptor, and Human Epidermal Growth Factor Receptor 2 (HER2) mRNA levels]. Genomic information was correlated with clinical and pathologic characteristics and Oncotype DX® Recurrence Score® (RS). One hundred Patients were enrolled, 1 not evaluable by BluePrint. The median age was 60 years (range 22–98), and eighty-four (84 %) patients had stage I or II disease. High Risk MammaPrint was present in 66 % of patients and in 52 % of patients with stage I disease. High Risk MammaPrint was associated with young age (p = 0.02), high grade (p < 0.0001), HER2 expression (p = 0.016), and triple-negative phenotype (p < 0.001). Sixty-four tumors (65 %) were Luminal type (47 % of these were classified as High Risk), 26 (26 %) were Basal type, and 9 (9 %) HER2 type. Twenty-two cancers were triple negative (TN) by IHC and 19 (90 %) Basal type. Among the 15 tumors HER2 positive by IHC/FISH, 8 (53 %) were HER2 type by BluePrint. Eleven tumors with ER expression of 1–9 % were ER negative by TargetPrint and none of these was Luminal type. None of the seven tumors HER2 positive by IHC/FISH but negative by TargetPrint was HER type. RS results were available in 29 patients: two had High Risk both by RS and MammaPrint; eight had intermediate RS, with four High Risk by MammaPrint; 19 had a low RS, with eight High Risk by MammaPrint. AAF with stage I to III breast cancer often present with High Risk disease. Molecular heterogeneity is present within TN, HER2-positive, and ER-positive breast cancer. RS and MammaPrint offer different prognostic information.


Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65(1):5–29
Wheeler SB, Reeder-Hayes KE, Carey LA (2013) Disparities in breast cancer treatment and outcomes: biological, social, and health system determinants and opportunities for research. Oncologist 18(9):986–993
Albain KS et al (2009) Racial disparities in cancer survival among randomized clinical trials patients of the Southwest Oncology Group. J Natl Cancer Inst 101(14):984–992
Wojcik BE, Spinks MK, Optenberg SA (1998) Breast carcinoma survival analysis for African American and white women in an equal-access health care system. Cancer 82(7):1310–1318
O’Brien KM et al (2010) Intrinsic breast tumor subtypes, race, and long-term survival in the Carolina Breast Cancer Study. Clin Cancer Res 16(24):6100–6110
Perou CM et al (2000) Molecular portraits of human breast tumours. Nature 406(6797):747–752
Sorlie T et al (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98(19):10869–10874
Sotiriou C, Pusztai L (2009) Gene-expression signatures in breast cancer. N Engl J Med 360(8):790–800
Carey LA et al (2006) Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295(21):2492–2502
Harris LN et al (2016) Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol 34(10):1134–1150
Sapino A et al (2014) MammaPrint molecular diagnostics on formalin-fixed, paraffin-embedded tissue. J Mol Diagn 16(2):190–197
Glas AM et al (2006) Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genom 7:278
Buyse M et al (2006) Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst 98(17):1183–1192
Krijgsman O et al (2012) A diagnostic gene profile for molecular subtyping of breast cancer associated with treatment response. Breast Cancer Res Treat 133(1):37–47
Bayraktar S et al (2014) Molecular subtyping predicts pathologic tumor response in early-stage breast cancer treated with neoadjuvant docetaxel plus capecitabine with or without trastuzumab chemotherapy. Med Oncol 31(10):163
Keenan T et al (2015) Comparison of the genomic landscape between primary breast cancer in African American versus white women and the association of racial differences with tumor recurrence. J Clin Oncol 33(31):3621–3627
Sweeney C et al (2014) Intrinsic subtypes from PAM50 gene expression assay in a population-based breast cancer cohort: differences by age, race, and tumor characteristics. Cancer Epidemiol Biomarkers Prev 23(5):714–724
Huo D et al (2009) Population differences in breast cancer: survey in indigenous African women reveals over-representation of triple-negative breast cancer. J Clin Oncol 27(27):4515–4521
van de Vijver MJ et al (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347(25):1999–2009
Mook S et al (2010) The 70-gene prognosis signature predicts early metastasis in breast cancer patients between 55 and 70 years of age. Ann Oncol 21(4):717–722
Drukker CA et al (2013) A prospective evaluation of a breast cancer prognosis signature in the observational RASTER study. Int J Cancer 133(4):929–936
Cardoso F, van’t Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, Pierga J-Y, Brain E, Causeret S, DeLorenzi M, Glas AM, Golfinopoulos V, Goulioti T, Knox S, Matos E, Meulemans B, Neijenhuis PA, Nitz U, Passalacqua R, Ravdin P, Rubio IT, Saghatchian M, Smilde TJ, Sotiriou C, Stork L, Straehle C, Thomas G, Thompson AM, van der Hoeven JM, Vuylsteke P, Bernards R, Tryfonidis K, Rutgers E, Piccart M (2016) 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med 375(8):717–729
Albain KS et al (2010) Potential biologic causes of the racial survival disparity in adjuvant trials of ER-positive breast cancer. J Clin Oncol (Meet Abstr) 28(15_suppl):511
Sparano JA et al (2012) Race and hormone receptor-positive breast cancer outcomes in a randomized chemotherapy trial. J Natl Cancer Inst 104(5):406–414
Prat A et al (2015) Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 24(Suppl 2):S26–S35
Perou CM (2011) Molecular stratification of triple-negative breast cancers. Oncologist 16(Suppl 1):61–70
Lehmann BD et al (2011) Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121(7):2750–2767
Carey LA et al (2016) Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol 34(6):542–549
Pogue-Geile KL et al (2015) Intrinsic subtypes, PIK3CA mutation, and the degree of benefit from adjuvant trastuzumab in the NSABP B-31 trial. J Clin Oncol 33(12):1340–1347
Iwamoto T et al (2012) Estrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1–10 % ER-positive by immunohistochemistry. J Clin Oncol 30(7):729–734
Hammond ME et al (2010) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 28(16):2784–2795
Wolff AC et al (2013) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 31(31):3997–4013
Clough K et al (2012) Abstract P6-07-03: risk classification of early stage breast cancer as assessed by MammaPrint and Oncotype DX genomic assays. Cancer Res 72(24 Supplement):P6-07-03
Bartlett JM et al (2016) Comparing breast cancer multiparameter tests in the OPTIMA prelim trial: no test is more equal than the others. J Natl Cancer Inst 108(9)
Curtis C et al (2012) The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 486(7403):346–352
Acknowledgments
MammaPrint, BluePrint, and TargetPrint tests were provided by Agendia. The biostatistical support was provided by the Georgetown-Howard Universities Center for Clinical and Translational Science (GHUCCTS). This study was supported in part by The Safeway Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Rights and permissions
About this article
Cite this article
Nunes, R.A., Wray, L., Mete, M. et al. Genomic profiling of breast cancer in African-American women using MammaPrint. Breast Cancer Res Treat 159, 481–488 (2016). https://doi.org/10.1007/s10549-016-3949-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-016-3949-y