Abstract
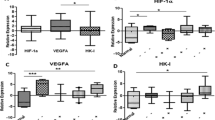
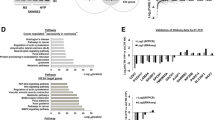
Numerous microarray-based gene expression studies performed on several types of solid tumors revealed significant changes in key genes involved in progression and regulation of the cell cycle, including AURKA that is known to be overexpressed in many types of human malignancies. Tumor hypoxia is associated with poor prognosis in several cancer types, including breast cancer (BC). Since hypoxia is a condition that influences the expression of many genes involved in tumorigenesis, proliferation, and cell cycle regulation, we performed a microarray-based gene expression analysis in order to identify differentially expressed genes in BC cell lines exposed to hypoxia. This analysis showed that hypoxia induces a down-regulation of AURKA expression. Although hypoxia is a tumor feature, the molecular mechanisms that regulate AURKA expression in response to hypoxia in BC are still unknown. For the first time, we demonstrated that HIF-1 activation downstream of hypoxia could drive AURKA down-regulation in BC cells. In fact, we found that siRNA-mediated knockdown of HIF-1α significantly reduces the AURKA down-regulation in BC cells under hypoxia. The aim of our study was to obtain new insights into AURKA transcriptional regulation in hypoxic conditions. Luciferase reporter assays showed a reduction of AURKA promoter activity in hypoxia. Unlike the previous findings, we hypothesize a new possible mechanism where HIF-1, rather than inducing transcriptional activation, could promote the AURKA down-regulation via its binding to hypoxia-responsive elements into the proximal region of the AURKA promoter. The present study shows that hypoxia directly links HIF-1 with AURKA expression, suggesting a possible pathophysiological role of this new pathway in BC and confirming HIF-1 as an important player linking an environmental signal to the AURKA promoter. Since AURKA down-regulation overrides the estrogen-mediated growth and chemoresistance in BC cells, these findings could be important for the development of new possible therapies against BC.







Similar content being viewed by others
References
Lech R, Przemyslaw O (2011) Epidemiological models for breast cancer risk estimation. Ginekol Pol 82(6):451–454
Alvarez RH, Valero V, Hortobagyi GN (2010) Emerging targeted therapies for breast cancer. J Clin Oncol 28(20):3366–3379. doi:10.1200/JCO.2009.25.4011
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98(19):10869–10874. doi:10.1073/pnas.191367098
Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D (2002) Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell 13(6):1977–2000. doi:10.1091/mbc.02-02-0030
Liu J, Campen A, Huang S, Peng SB, Ye X, Palakal M, Dunker AK, Xia Y, Li S (2008) Identification of a gene signature in cell cycle pathway for breast cancer prognosis using gene expression profiling data. BMC Med Genomics 1:39. doi:10.1186/1755-8794-1-39
Golias CH, Charalabopoulos A, Charalabopoulos K (2004) Cell proliferation and cell cycle control: a mini review. Int J Clin Pract 58(12):1134–1141
Williams GH, Stoeber K (2012) The cell cycle and cancer. J Pathol 226(2):352–364. doi:10.1002/path.3022
Fu J, Bian M, Jiang Q, Zhang C (2007) Roles of aurora kinases in mitosis and tumorigenesis. Mol Cancer Res 5(1):1–10. doi:10.1158/1541-7786.MCR-06-0208
Vader G, Lens SM (2008) The aurora kinase family in cell division and cancer. Biochim Biophys Acta 1786(1):60–72. doi:10.1016/j.bbcan.2008.07.003
Barr AR, Gergely F (2007) Aurora-A: the maker and breaker of spindle poles. J Cell Sci 120(Pt 17):2987–2996. doi:10.1242/jcs.013136
El-Rifai W, Frierson HF Jr, Harper JC, Powell SM, Knuutila S (2001) Expression profiling of gastric adenocarcinoma using cDNA array. Int J Cancer 92(6):832–838. doi:10.1002/ijc.1264
Tanaka T, Kimura M, Matsunaga K, Fukada D, Mori H, Okano Y (1999) Centrosomal kinase AIK1 is overexpressed in invasive ductal carcinoma of the breast. Cancer Res 59(9):2041–2044
Sen S, Zhou H, Zhang RD, Yoon DS, Vakar-Lopez F, Ito S, Jiang F, Johnston D, Grossman HB, Ruifrok AC, Katz RL, Brinkley W, Czerniak B (2002) Amplification/overexpression of a mitotic kinase gene in human bladder cancer. J Natl Cancer Inst 94(17):1320–1329
Klein A, Flugel D, Kietzmann T (2008) Transcriptional regulation of serine/threonine kinase-15 (STK15) expression by hypoxia and HIF-1. Mol Biol Cell 19(9):3667–3675. doi:10.1091/mbc.E08-01-0042
Wu CC, Yang TY, Yu CT, Phan L, Ivan C, Sood AK, Hsu SL, Lee MH (2012) p53 negatively regulates aurora A via both transcriptional and posttranslational regulation. Cell Cycle 11(18):3433–3442. doi:10.4161/cc.21732
Lehman NL, O’Donnell JP, Whiteley LJ, Stapp RT, Lehman TD, Roszka KM, Schultz LR, Williams CJ, Mikkelsen T, Brown SL, Ecsedy JA, Poisson LM (2012) Aurora A is differentially expressed in gliomas, is associated with patient survival in glioblastoma and is a potential chemotherapeutic target in gliomas. Cell Cycle 11(3):489–502. doi:10.4161/cc.11.3.18996
Agnese V, Bazan V, Fiorentino FP, Fanale D, Badalamenti G, Colucci G, Adamo V, Santini D, Russo A (2007) The role of aurora-A inhibitors in cancer therapy. Ann Oncol 18(Suppl 6):47–52. doi:10.1093/annonc/mdm224
Rademakers SE, Span PN, Kaanders JH, Sweep FC, van der Kogel AJ, Bussink J (2008) Molecular aspects of tumour hypoxia. Mol Oncol 2(1):41–53. doi:10.1016/j.molonc.2008.03.006
Brown JM (2002) Tumor microenvironment and the response to anticancer therapy. Cancer Biol Ther 1(5):453–458
Semenza G (2002) Signal transduction to hypoxia-inducible factor 1. Biochem Pharmacol 64(5–6):993–998
Knowles HJ, Harris AL (2001) Hypoxia and oxidative stress in breast cancer. Hypoxia and tumourigenesis. Breast Cancer Res 3(5):318–322
Wenger A, Kowalewski N, Stahl A, Mehlhorn AT, Schmal H, Stark GB, Finkenzeller G (2005) Development and characterization of a spheroidal coculture model of endothelial cells and fibroblasts for improving angiogenesis in tissue engineering. Cells Tissues Organs 181(2):80–88. doi:10.1159/000091097
Schwab LP, Peacock DL, Majumdar D, Ingels JF, Jensen LC, Smith KD, Cushing RC, Seagroves TN (2012) Hypoxia-inducible factor 1α promotes primary tumor growth and tumor-initiating cell activity in breast cancer. Breast Cancer Res 14(1):R6. doi:10.1186/bcr3087
Pugh CW, Gleadle J, Maxwell PH (2001) Hypoxia and oxidative stress in breast cancer. Hypoxia signalling pathways. Breast Cancer Res 3(5):313–317
Kimbro KS, Simons JW (2006) Hypoxia-inducible factor-1 in human breast and prostate cancer. Endocr Relat Cancer 13(3):739–749. doi:10.1677/erc.1.00728
Vaupel P (2004) The role of hypoxia-induced factors in tumor progression. Oncologist 9(suppl 5):10–17. doi:10.1634/theoncologist.9-90005-10
Peurala E, Koivunen P, Bloigu R, Haapasaari KM, Jukkola-Vuorinen A (2012) Expressions of individual PHDs associate with good prognostic factors and increased proliferation in breast cancer patients. Breast Cancer Res Treat 133(1):179–188. doi:10.1007/s10549-011-1750-5
Kaelin WG Jr (2003) The von Hippel-Lindau gene, kidney cancer, and oxygen sensing. J Am Soc Nephrol 14(11):2703–2711
Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL (2005) Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 105(2):659–669. doi:10.1182/blood-2004-07-2958
Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC (2003) Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol 23(24):9361–9374
Giatromanolaki A, Harris AL (2001) Tumour hypoxia, hypoxia signaling pathways and hypoxia inducible factor expression in human cancer. Anticancer Res 21(6):4317–4324
Yamamoto Y, Ibusuki M, Okumura Y, Kawasoe T, Kai K, Iyama K, Iwase H (2008) Hypoxia-inducible factor 1α is closely linked to an aggressive phenotype in breast cancer. Breast Cancer Res Treat 110(3):465–475. doi:10.1007/s10549-007-9742-1
Chen KF, Lai YY, Sun HS, Tsai SJ (2005) Transcriptional repression of human cad gene by hypoxia inducible factor-1α. Nucl Acids Res 33(16):5190–5198. doi:10.1093/nar/gki839
Feige E, Yokoyama S, Levy C, Khaled M, Igras V, Lin RJ, Lee S, Widlund HR, Granter SR, Kung AL, Fisher DE (2011) Hypoxia-induced transcriptional repression of the melanoma-associated oncogene MITF. Proc Natl Acad Sci USA 108(43):E924–E933. doi:10.1073/pnas.1106351108
Ryu K, Park C, Lee Y (2011) Hypoxia-inducible factor 1 alpha represses the transcription of the estrogen receptor alpha gene in human breast cancer cells. Biochem Biophys Res Commun 407(4):831–836. doi:10.1016/j.bbrc.2011.03.119
Federico M, Symonds CE, Bagella L, Rizzolio F, Fanale D, Russo A, Giordano A (2010) R-Roscovitine (seliciclib) prevents DNA damage-induced cyclin A1 upregulation and hinders non-homologous end-joining (NHEJ) DNA repair. Mol Cancer 9:208. doi:10.1186/1476-4598-9-208
Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20(3):307–315. doi:10.1093/bioinformatics/btg405
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4(2):249–264. doi:10.1093/biostatistics/4.2.249
Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3, Article 3. doi:10.2202/1544-6115.1027
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125(1–2):279–284
Wettenhall JM, Simpson KM, Satterley K, Smyth GK (2006) affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics 22(7):897–899. doi:10.1093/bioinformatics/btl025
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucl Acids Res 28(1):27–30
DeSantis C, Siegel R, Bandi P, Jemal A (2011) Breast cancer statistics. CA Cancer J Clin 61(6):409–418. doi:10.3322/caac.20134
van der Groep P, van Diest PJ, Smolders YH, Ausems MG, van der Luijt RB, Menko FH, Bart J, de Vries EG, van der Wall E (2013) HIF-1alpha overexpression in ductal carcinoma in situ of the breast in BRCA1 and BRCA2 mutation carriers. PLoS ONE 8(2):e56055. doi:10.1371/journal.pone.0056055
Favaro E, Lord S, Harris AL, Buffa FM (2011) Gene expression and hypoxia in breast cancer. Genome Med 3(8):55. doi:10.1186/gm271
Kunz M, Ibrahim SM (2003) Molecular responses to hypoxia in tumor cells. Mol Cancer 2:23
Lens SM, Voest EE, Medema RH (2010) Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer 10(12):825–841. doi:10.1038/nrc2964
Kitajima S, Kudo Y, Ogawa I, Tatsuka M, Kawai H, Pagano M, Takata T (2007) Constitutive phosphorylation of aurora-A on ser51 induces its stabilization and consequent overexpression in cancer. PLoS ONE 2(9):e944. doi:10.1371/journal.pone.0000944
Katayama H, Brinkley WR, Sen S (2003) The aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev 22(4):451–464
Wang LH, Xiang J, Yan M, Zhang Y, Zhao Y, Yue CF, Xu J, Zheng FM, Chen JN, Kang Z, Chen TS, Xing D, Liu Q (2010) The mitotic kinase aurora-A induces mammary cell migration and breast cancer metastasis by activating the Cofilin-F-actin pathway. Cancer Res 70(22):9118–9128. doi:10.1158/0008-5472.CAN-10-1246
Yang H, He L, Kruk P, Nicosia SV, Cheng JQ (2006) Aurora-A induces cell survival and chemoresistance by activation of Akt through a p53-dependent manner in ovarian cancer cells. Int J Cancer 119(10):2304–2312. doi:10.1002/ijc.22154
Xu J, Li H, Wang B, Xu Y, Yang J, Zhang X, Harten SK, Shukla D, Maxwell PH, Pei D, Esteban MA (2010) VHL inactivation induces HEF1 and aurora kinase A. J Am Soc Nephrol 21(12):2041–2046. doi:10.1681/ASN.2010040345
Lee HH, Zhu Y, Govindasamy KM, Gopalan G (2008) Downregulation of aurora-A overrides estrogen-mediated growth and chemoresistance in breast cancer cells. Endocr Relat Cancer 15(3):765–775. doi:10.1677/ERC-07-0213
Conflict of interest
The authors declare no conflict of interest.
Ethical standards
The authors declare that the experiments comply with the current laws.
Author information
Authors and Affiliations
Corresponding author
Additional information
Daniele Fanale and Viviana Bazan have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Pattern of common differentially expressed genes in breast cancer cell lines in response to hypoxia (XLS 83 kb)
Table S2
DEGs involved in cell cycle regulation pathway (XLS 28 kb)
Table S3
DEGs involved in HIF-1a network (XLS 24 kb)
Table S4
DEGs involved in AURKA signaling pathway (XLS 23 kb)
Rights and permissions
About this article
Cite this article
Fanale, D., Bazan, V., Corsini, L.R. et al. HIF-1 is involved in the negative regulation of AURKA expression in breast cancer cell lines under hypoxic conditions. Breast Cancer Res Treat 140, 505–517 (2013). https://doi.org/10.1007/s10549-013-2649-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-013-2649-0