Abstract
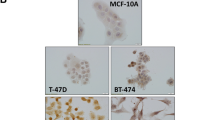
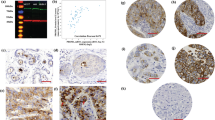
Drosha is a protein that plays a key role in the biogenesis of microRNAs which are well known to be deranged in human breast cancer (BC). The purpose of the current study was to assess the biological and prognostic value of Drosha protein expression in BC. Drosha protein expression was assessed immunohistochemically in two sets of BC: (1) full-face sections of selected BC series with distinct stages of tumour progression (Normal parenchymal cells, ductal carcinoma in situ (DCIS), primary invasive BC and nodal metastases) to evaluate its differential expression, (2) tissue microarray comprising a large and well-characterised series of unselected clinically annotated invasive BC to investigate its correlation with clinicopathological features and patient outcome. A gradual loss of Drosha cytoplasmic expression was observed along tumour progression from DCIS, to invasive and to metastatic cancer cells. In invasive BC, loss of Drosha cytoplasmic expression was associated with BRCA1 and ER expression and with shorter BC specific survival (BCSS), disease free interval (DFI) and distant metastasis free interval (DMFI). This correlation was maintained in ER negative, HER2 negative, triple negative and LN negative cases. Moreover, loss of cytoplasmic Drosha was predictive of better response to chemotherapy and endocrine therapy. This study provides evidence that Drosha protein potentially plays an important role in BC progression and assessment of its expression provides an independent predictor of patient outcome. These observations provide further evidence that alterations in miRNA regulation influence tumour behaviour.






Similar content being viewed by others
References
Brennecke J et al (2003) Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113(1):25–36
Miska EA, Miska EA (2005) How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev 15(5):563–568
Reinhart BJ et al (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403(6772):901–906
Xu P et al (2003) The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol 13(9):790–795
Lee Y et al (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23(20):4051–4060
Denli AM et al (2004) Processing of primary microRNAs by the microprocessor complex. Nature 432(7014):231–235
Gregory RI et al (2004) The Microprocessor complex mediates the genesis of microRNAs. Nature 432(7014):235–240
Lee Y et al (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425(6956):415–419
Lund E et al (2004) Nuclear export of microRNA precursors. Science 303(5654):95–98
Hutvagner G et al (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293(5531):834–838
Khoshnaw SM et al (2009) MicroRNA involvement in the pathogenesis and management of breast cancer. J Clin Pathol 62(5):422–428
Kawai S, Amano A (2012) BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J Cell Biol 197(2):201–208
Suzuki HI et al (2009) Modulation of microRNA processing by p53. Nature 460(7254):529–533
Davis BN et al (2008) SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454(7200):56–61
Yamagata K et al (2009) Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell 36(2):340–347
Dedes KJ et al (2011) Down-regulation of the miRNA master regulators Drosha and Dicer is associated with specific subgroups of breast cancer. Eur J Cancer 47(1):138–150
Merritt WM et al (2008) Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med 359(25):2641–2650
Shu GS, Yang ZL, Liu DC (2012) Immunohistochemical study of Dicer and Drosha expression in the benign and malignant lesions of gallbladder and their clinicopathological significances. Pathol Res Pract 208(7):392–397
Rakha EA et al (2005) Morphological and immunophenotypic analysis of breast carcinomas with basal and myoepithelial differentiation. J Pathol 208(4):495–506
Rakha EA et al (2007) Prognostic markers in triple-negative breast cancer. Cancer 109(1):25–32
Rakha EA et al (2005) Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Mod Pathol 18(10):1295–1304
Abd El-Rehim DM et al (2005) High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer 116(3):340–350
Rakha EA et al (2009) Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res 15(7):2302–2310
Galea MH et al (1992) The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat 22(3):207–219
Abd El-Rehim DM et al (2004) Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma. Br J Cancer 91(8):1532–1542
Rakha EA et al (2010) Clinical and biological significance of E-cadherin protein expression in invasive lobular carcinoma of the breast. Am J Surg Pathol 34(10):1472–1479
Kononen J et al (1998) Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 4(7):844–847
Novocastra, Novolink Polymer detection system customer guidance booklet for immunohistochemistry procedure, Leica Microsystems, www.leica-microsystems.com
McCarty KS Jr et al (1985) Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med 109(8):716–721
McShane LM et al (2006) REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat 100(2):229–235
Wu H et al (2000) Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J Biol Chem 275(47):36957–36965
Shapiro JS et al (2012) Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA 18(7):1338–1346
Passon N et al (2012) Expression of Dicer and Drosha in triple-negative breast cancer. J Clin Pathol 65(4):320–326
Yan M et al (2012) Dysregulated expression of Dicer and Drosha in breast cancer. Pathol Oncol Res 18(2):343–348
Lin RJ et al (2010) microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res 70(20):7841–7850
Somasundaram K (2003) Breast cancer gene 1 (BRCA1): role in cell cycle regulation and DNA repair–perhaps through transcription. J Cell Biochem 88(6):1084–1091
Paull TT et al (2001) Direct DNA binding by Brca1. Proc Natl Acad Sci USA 98(11):6086–6091
Vaksman O et al (2012) Argonaute, Dicer, and Drosha are up-regulated along tumor progression in serous ovarian carcinoma. Hum Pathol 43(11):2062–2069
Rakha EA et al (2007) Breast carcinoma with basal differentiation: a proposal for pathology definition based on basal cytokeratin expression. Histopathology 50(4):434–438
Conflicts of interest
The authors have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khoshnaw, S.M., Rakha, E.A., Abdel-Fatah, T. et al. The microRNA maturation regulator Drosha is an independent predictor of outcome in breast cancer patients. Breast Cancer Res Treat 137, 139–153 (2013). https://doi.org/10.1007/s10549-012-2358-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-012-2358-0