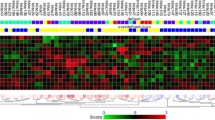
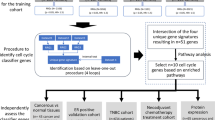
Abstract
Prognosis of early beast cancer is heterogeneous. Today, no histoclinical or biological factor predictive for clinical outcome after adjuvant anthracycline-based chemotherapy (CT) has been validated and introduced in routine use. Using DNA microarrays, we searched for a gene expression signature associated with metastatic relapse after adjuvant anthracycline-based CT without taxane. We profiled a multicentric series of 595 breast cancers including 498 treated with such adjuvant CT. The identification of the prognostic signature was done using a metagene-based supervised approach in a learning set of 323 patients. The signature was then tested on an independent validation set comprising 175 similarly treated patients, 128 of them from the PACS01 prospective clinical trial. We identified a 3-metagene predictor of metastatic relapse in the learning set, and confirmed its independent prognostic impact in the validation set. In multivariate analysis, the predictor outperformed the individual current prognostic factors, as well as the Nottingham Prognostic Index-based classifier, both in the learning and the validation sets, and added independent prognostic information. Among the patients treated with adjuvant anthracycline-based CT, with a median follow-up of 68 months, the 5-year metastasis-free survival was 82% in the “good-prognosis” group and 56% in the “poor-prognosis” group. Our predictor refines the prediction of metastasis-free survival after adjuvant anthracycline-based CT and might help tailoring adjuvant CT regimens.


Similar content being viewed by others
Abbreviations
- CT:
-
Chemotherapy
- IPC:
-
Institut Paoli-Calmettes
- CLB:
-
Centre Léon Bérard
- IB:
-
Institut Bergonié
- SBR:
-
Scarff-Bloom-Richardson
- IHC:
-
Immunohistochemistry
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- FEC:
-
5-Fluoro-uracile + epirubicine + cyclophosphamide
- PCR:
-
Polymerase chain reaction
- MFS:
-
Metastasis-free survival
- HR:
-
Hazard ratio
References
EBCTCI Group (2005) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomized trials. Lancet 365:1687–1717
Piccart-Gebhart MJ, Procter M, Leyland-Jones B et al (2005) Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med 353:1659–1672
Gonzalez-Angulo AM, Hortobagyi GN, Esteva FJI (2006) Adjuvant therapy with trastuzumab for HER-2/neu-positive breast cancer. Oncologist 11:857–867
Romond EH, Perez EA, Bryant J et al (2005) Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med 353:1673–1684
Sparano JA, Hortobagyi GN, Gralow JR, Perez EA, Comis RLI (2010) Recommendations for research priorities in breast cancer by the coalition of cancer cooperative groups scientific leadership council: systemic therapy and therapeutic individualization. Breast Cancer Res Treat 119:511–527
Goldhirsch A, Glick JH, Gelber RD, Coates AS, Senn HJI (2001) Meeting highlights: international consensus panel on the treatment of primary breast cancer. Seventh international conference on adjuvant therapy of primary breast cancer. J Clin Oncol 19:3817–3827
Colozza M, de Azambuja E, Cardoso F, Bernard C, Piccart MJI (2006) Breast cancer: achievements in adjuvant systemic therapies in the pre-genomic era. Oncologist 11:111–125
Piccart MJ, de Valeriola D, Dal Lago L et al (2005) Adjuvant chemotherapy in 2005: standards and beyond. Breast 14:439–445
Buzdar AU, Singletary SE, Valero V et al (2002) Evaluation of paclitaxel in adjuvant chemotherapy for patients with operable breast cancer: preliminary data of a prospective randomized trial. Clin Cancer Res 8:1073–1079
Henderson IC, Berry DA, Demetri GD et al (2003) Improved outcomes from adding sequential paclitaxel but not from escalating doxorubicin dose in an adjuvant chemotherapy regimen for patients with node-positive primary breast cancer. J Clin Oncol 21:976–983
Mamounas EP, Bryant J, Lembersky B et al (2005) Paclitaxel after doxorubicin plus cyclophosphamide as adjuvant chemotherapy for node-positive breast cancer: results from NSABP B-28. J Clin Oncol 23:3686–3696
Martin M, Pienkowski T, Mackey J et al (2005) Adjuvant docetaxel for node-positive breast cancer. N Engl J Med 352:2302–2313
Roche H, Fumoleau P, Spielmann M et al (2006) Sequential adjuvant epirubicin-based and docetaxel chemotherapy for node-positive breast cancer patients: the PACS01 trial. J Clin Oncol 24:5664–5671
Hayes DFI (2005) Prognostic and predictive factors revisited. Breast 14:493–499
Hayes DFI (2009) Is there a standard type and duration of adjuvant chemotherapy for early stage breast cancer? Breast 18(Suppl 3):S131–S134
Bertucci F, Finetti P, Cervera N et al (2006) Gene expression profiling and clinical outcome in breast cancer. Omics 10:429–443
Sorlie T, Perou CM, Tibshirani R et al (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98:10869–10874
Sorlie T, Tibshirani R, Parker J et al (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100:8418–8423
van de Vijver MJ, He YD, van’t Veer LJ et al (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347:1999–2009
van ‘t Veer LJ, Dai H, van de Vijver MJ et al (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530–536
Wang Y, Klijn JG, Zhang Y et al (2005) Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365:671–679
Foekens JA, Atkins D, Zhang Y et al (2006) Multicenter validation of a gene expression-based prognostic signature in lymph node-negative primary breast cancer. J Clin Oncol 24:1665–1671
Buyse M, Loi S, van’t Veer L et al (2006) Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst 98:1183–1192
Desmedt C, Piette F, Loi S et al (2007) Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res 13:3207–3214
Sotiriou C, Wirapati P, Loi S et al (2006) Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 98:262–272
Ivshina AV, George J, Senko O et al (2006) Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res 66:10292–10301
Miller LD, Smeds J, George J et al (2005) An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA 102:13550–13555
Naderi A, Teschendorff AE, Barbosa-Morais NL et al (2007) A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene 26:1507–1516
Ma XJ, Wang Z, Ryan PD et al (2004) A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell 5:607–616
Paik S, Shak S, Tang G et al (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351:2817–2826
Oh DS, Troester MA, Usary J et al (2006) Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J Clin Oncol 24:1656–1664
Loi S, Haibe-Kains B, Desmedt C et al (2007) Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol 25:1239–1246
Teschendorff AE, Naderi A, Barbosa-Morais NL et al (2006) A consensus prognostic gene expression classifier for ER positive breast cancer. Genome Biol 7:R101
Sotiriou C, Piccart MJI (2007) Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer 7:545–553
Ross JS, Hatzis C, Symmans WF, Pusztai L, Hortobagyi GNI (2008) Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist 13:477–493
Ayers M, Symmans WF, Stec J et al (2004) Gene expression profiles predict complete pathologic response to neoadjuvant paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide chemotherapy in breast cancer. J Clin Oncol 22:2284–2293
Bertucci F, Finetti P, Rougemont J et al (2004) Gene expression profiling for molecular characterization of inflammatory breast cancer and prediction of response to chemotherapy. Cancer Res 64:8558–8565
Chang JC, Wooten EC, Tsimelzon A et al (2003) Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet 362:362–369
Hannemann J, Oosterkamp HM, Bosch CA et al (2005) Changes in gene expression associated with response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol 23:3331–3342
Bonnefoi H, Potti A, Delorenzi M et al (2007) Validation of gene signatures that predict the response of breast cancer to neoadjuvant chemotherapy: a substudy of the EORTC 10994/BIG 00–01 clinical trial. Lancet Oncol 8:1071–1078
Folgueira MA, Carraro DM, Brentani H et al (2005) Gene expression profile associated with response to doxorubicin-based therapy in breast cancer. Clin Cancer Res 11:7434–7443
Hess KR, Anderson K, Symmans WF et al (2006) Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol 24:4236–4244
Iwao-Koizumi K, Matoba R, Ueno N et al (2005) Prediction of docetaxel response in human breast cancer by gene expression profiling. J Clin Oncol 23:422–431
Farmer P, Bonnefoi H, Anderle P et al (2009) A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med 15:68–74
Bertucci F, Houlgatte R, Benziane A et al (2000) Gene expression profiling of primary breast carcinomas using arrays of candidate genes. Hum Mol Genet 9:2981–2991
Bertucci F, Nasser V, Granjeaud S et al (2002) Gene expression profiles of poor-prognosis primary breast cancer correlate with survival. Hum Mol Genet 11:863–872
Specht K, Harbeck N, Smida J et al (2009) Expression profiling identifies genes that predict recurrence of breast cancer after adjuvant CMF-based chemotherapy. Breast Cancer Res Treat 118:45–56
Campone M, Campion L, Roche H et al (2008) Prediction of metastatic relapse in node-positive breast cancer: establishment of a clinicogenomic model after FEC100 adjuvant regimen. Breast Cancer Res Treat 109:491–501
Pawitan Y, Bjohle J, Amler L et al (2005) Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res 7:R953–R964
Jezequel P, Campone M, Roche H et al (2009) A 38-gene expression signature to predict metastasis risk in node-positive breast cancer after systemic adjuvant chemotherapy: a genomic substudy of PACS01 clinical trial. Breast Cancer Res Treat 116:509–520
Bertucci F, Borie N, Ginestier C et al (2004) Identification and validation of an ERBB2 gene expression signature in breast cancers. Oncogene 23:2564–2575
Vey N, Mozziconacci MJ, Groulet-Martinec A et al (2004) Identification of new classes among acute myelogenous leukaemias with normal karyotype using gene expression profiling. Oncogene 23:9381–9391
Bertucci F, Orsetti B, Negre V et al (2008) Lobular and ductal carcinomas of the breast have distinct genomic and expression profiles. Oncogene 27:5359–5372
Kaplan EL, Meier PI (1958) Non-parametric estimation for incomplete observation. J Am Stat Assoc 53:457–481
Cox DRI (1972) Regression models and life table. J R Stat Soc [B] 34:187–220
Todd JH, Dowle C, Williams MR et al (1987) Confirmation of a prognostic index in primary breast cancer. Br J Cancer 56:489–492
Gruvberger S, Ringner M, Chen Y et al (2001) Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res 61:5979–5984
Perou CM, Sorlie T, Eisen MB et al (2000) Molecular portraits of human breast tumours. Nature 406:747–752
West M, Blanchette C, Dressman H et al (2001) Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci USA 98:11462–11467
Bertucci F, Finetti P, Cervera N et al (2006) Gene expression profiling shows medullary breast cancer is a subgroup of basal breast cancers. Cancer Res 66:4636–4644
Desmedt C, Haibe-Kains B, Wirapati P et al (2008) Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res 14:5158–5165
Dowsett M, Goldhirsch A, Hayes DF et al (2007) International Web-based consultation on priorities for translational breast cancer research. Breast Cancer Res 9:R81
Bonadonna G, Valagussa P, Moliterni A, Zambetti M, Brambilla CI (1995) Adjuvant cyclophosphamide, methotrexate, and fluorouracil in node-positive breast cancer: the results of 20 years of follow-up. N Engl J Med 332:901–906
Demicheli R, Miceli R, Brambilla C et al (1999) Comparative analysis of breast cancer recurrence risk for patients receiving or not receiving adjuvant cyclophosphamide, methotrexate, fluorouracil (CMF). Data supporting the occurrence of ‘cures’. Breast Cancer Res Treat 53:209–215
Bertucci F, Ferrero JM, Bachelot T et aI (2009) SA02 trial: a genomics-based prospective study of adjuvant chemotherapy in node-positive early breast cancer with “good-prognosis signature”. SABCS 2009
Piccart-Gebhart MJI (2005) Moving away from the “one shoe fits all” strategy: the key to future progress in chemotherapy. J Clin Oncol 23:1611–1613
Acknowledgments
We are grateful to L. Xerri and C. Mawas for encouragements. We thank F. Bonnet, S. Mathoulin-Pélissier, V. Brouste for their technical assistance and S. Deraco for his help in data analysis. This study was supported in part by Inserm, Institut Paoli-Calmettes, and grants from Programme Hospitalier de Recherche Clinique 2001 (PHRC N° 24-01 FB) and 2003 (N° 24-01 FB), Ligue Nationale Contre le Cancer (label DB), Association pour la Recherche sur le Cancer, Fédération Nationale des Centres de Lutte Contre le Cancer and Institut National du Cancer (ACI2004, PL2006, recherche translationnelle). The Biological Resource Centres in Oncology at Institut Paoli-Calmettes and Centre Léon Bérard were supported by grants “Tumorothèques” from the French Ministry of Health and INCa, by grants “Collections 2003” from Inserm and the French Ministry of Research, and by grants “CEBS 2006” from the Agence Nationale de la Recherche (ANR).
Conflict of interest
Ipsogen employees: N Borie, JM Le Doussal, S Debono, A Martinec, F Hermitte, V Fert. Ipsogen shareholders: S Debono, F Hermitte, V Fert. The other authors declare they have no competing interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10549_2010_1003_MOESM1_ESM.tif
Supplementary Fig. 1: Kaplan–Meier MFS for patients treated with adjuvant anthracycline-based chemotherapy. a MFS for all 498 patients. b–d MFS in the “good-prognosis group” or the “poor-prognosis group” defined using the genomic predictor; c MFS for all 498 patients; cMFS for 417 patients with N+ cancer; d MFS for 80 patients with N− cancer. HR means hazard ratio for metastasis in the “poor-prognosis group” as compared to the “good-prognosis group.” (TIFF 492 kb)
Rights and permissions
About this article
Cite this article
Bertucci, F., Borie, N., Roche, H. et al. Gene expression profile predicts outcome after anthracycline-based adjuvant chemotherapy in early breast cancer. Breast Cancer Res Treat 127, 363–373 (2011). https://doi.org/10.1007/s10549-010-1003-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-010-1003-z