Abstract
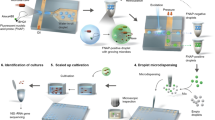
Peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) is a highly specific molecular method widely used for microbial identification. Nonetheless, and due to the detection limit of this technique, a time-consuming pre-enrichment step is typically required before identification. In here we have developed a lab-on-a-chip device to concentrate cell suspensions and speed up the identification process in yeasts. The PNA-FISH protocol was optimized to target Saccharomyces cerevisiae, a common yeast that is very relevant for several types of food industries. Then, several coin-sized microfluidic devices with different geometries were developed. Using Computational fluid dynamics (CFD), we modeled the hydrodynamics inside the microchannels and selected the most promising options. SU-8 structures were fabricated based on the selected designs and used to produce polydimethylsiloxane-based microchips by soft lithography. As a result, an integrated approach combining microfluidics and PNA-FISH for the rapid identification of S. cerevisiae was achieved. To improve fluid flow inside microchannels and the PNA-FISH labeling, oxygen plasma treatment was applied to the microfluidic devices and a new methodology to introduce the cell suspension and solutions into the microchannels was devised. A strong PNA-FISH signal was observed in cells trapped inside the microchannels, proving that the proposed methodology works as intended. The microfluidic designs and PNA-FISH procedure described in here should be easily adaptable for detection of other microorganisms of similar size.






Similar content being viewed by others
References
I. Abubakar, L. Irvine, C. Aldus, G. Wyatt, R. Fordham, S. Schelenz, L. Shepstone, A. Howe, M. Peck, P. Hunter, "A systematic review of the clinical, public health and cost-effectiveness of rapid diagnostic tests for the detection and identification of bacterial intestinal pathogens in faeces and food." health Technol. Assess. 11(36), 1–216 (2007)
C. Almeida, N.F. Azevedo, R. Fernandes, C.W. Keevil, M. Vieira, "fluorescence in situ hybridization method using a peptide nucleic acid probe for identification of Salmonella spp. in a broad spectrum of samples." Appl. Environ. Microbiol. 76(13), 4476–4485 (2010)
J. Alvankarian, A. Bahadorimehr, B.Y. Majlis, A pillar-based microfilter for isolation of white blood cells on elastomeric substrate. Biomicrofluidics 7(1), 014102 (2013)
R. Amann, B.M. Fuchs, Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nature Rev. Microbiol. 6(5), 339–348 (2008)
R.I. Amann, B. Zarda, D. Stahl, K. Schleifer, Identification of individual prokaryotic cells by using enzyme-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 58(9), 3007–3011 (1992)
J.R. Anderson, D.T. Chiu, R.J. Jackman, O. Cherniavskaya, J.C. McDonald, H. Wu, S.H. Whitesides, G.M. Whitesides, Fabrication of topologically complex three-dimensional microfluidic systems in PDMS by rapid prototyping. Anal. Chem. 72(14), 3158–3164 (2000)
I. Andorra, M. Monteiro, B. Esteve-Zarzoso, H. Albergaria, A. Mas, Analysis and direct quantification of Saccharomyces cerevisiae and Hanseniaspora guilliermondii populations during alcoholic fermentation by fluorescence in situ hybridization, flow cytometry and quantitative PCR. Food Microbiol. 28(8), 1483–1491 (2011)
ANSYS® Fluent, Release 12.0, getting started guide (ANSYS Inc., Canonsburg, 2009)
B. Bottari, D. Ercolini, M. Gatti, E. Neviani, Application of FISH technology for microbiological analysis: current state and prospects. Appl. Microbiol. Biotechnol. 73(3), 485–494 (2006)
L. Cerqueira, N.F. Azevedo, C. Almeida, T. Jardim, C.W. Keevil, M.J. Vieira, DNA mimics for the rapid identification of microorganisms by fluorescence in situ hybridization (FISH). Int. J. Mol. Sci. 9(10), 1944–1960 (2008)
L. Cerqueira, R.M. Fernandes, R.M. Ferreira, M. Oleastro, F. Carneiro, C. Brandão, P. Pimentel-Nunes, M. Dinis-Ribeiro, C. Figueiredo, C.W. Keevil, Validation of a fluorescence in situ hybridization method using peptide nucleic acid probes for detection of Helicobacter pylori clarithromycin resistance in gastric biopsy specimens. J. Clin. Microbiol. 51(6), 1887–1893 (2013)
R. Davidsson, B. Johansson, V. Passoth, M. Bengtsson, T. Laurell, J. Emnéus, Microfluidic biosensing systems part II. Monitoring the dynamic production of glucose and ethanol from microchip-immobilised yeast cells using enzymatic chemiluminescent μ-biosensors. Lab Chip 4(5), 488–494 (2004)
J.P. Devadhasan, S. Kim, J. An, Fish-on-a-chip: a sensitive detection microfluidic system for alzheimer's disease. J. Biomed. Sci. 18(1), 33 (2011)
H. Feldmann, Yeast: molecular and cell biology (John Wiley & Sons, Hoboken, 2011)
G. Gerdts, G. Luedke, FISH and chips: marine bacterial communities analyzed by flow cytometry based on microfluidics. J. Microbiol. Methods 64(2), 232–240 (2006)
J.R. Hart, Ethylenediaminetetraacetic acid and related chelating agents. Ullmann’s Encyclopedia of Industrial Chemistry (John Wiley & Sons, Hoboken, 2000)
J. Heo, S.Z. Hua, An overview of recent strategies in pathogen sensing. Sensors 9(6), 4483–4502 (2009)
H. Hillborg, J. Ankner, U.W. Gedde, G. Smith, H. Yasuda, K. Wikström, Crosslinked polydimethylsiloxane exposed to oxygen plasma studied by neutron reflectometry and other surface specific techniques. Polymer 41(18), 6851–6863 (2000)
S.M. Hong, S.H. Kim, J.H. Kim, H.I. Hwang, Hydrophilic surface modification of PDMS using atmospheric RF plasma. J. Phys. Conf. Ser. 34(1), 656–661 (IOP Publishing, 2006)
D. Huber, J. Rudolf, P. Ansari, B. Galler, M. Führer, C. Hasenhindl, S. Baumgartner, Effectiveness of natural and synthetic blocking reagents and their application for detecting food allergens in enzyme-linked immunosorbent assays. Anal. Bioanal. Chem. 394(2), 539–548 (2009)
Y.H. Hui, L.M. Nollet, F. Toldrá, Advances in food diagnostics (John Wiley & Sons, Hoboken, 2008)
R.I. Issa, Solution of the implicitly discretised fluid flow equations by operator-splitting. J. Comput. Phys. 62(1), 40–65 (1986)
E. Lagally, Microfluidics and nanotechnology: biosensing to the single molecule limit (CRC Press, Boca Raton, 2014)
B.P. Leonard, A stable and accurate convective modelling procedure based on quadratic upstream interpolation. Comput. Methods Appl. Mech. Eng. 19(1), 59–98 (1979)
P.C. Li, L. de Camprieu, J. Cai, M. Sangar, Transport, retention and fluorescent measurement of single biological cells studied in microfluidic chips. Lab Chip 4(3), 174–180 (2004)
H. Makamba, J.H. Kim, K. Lim, N. Park, J.H. Hahn, Surface modification of poly (dimethylsiloxane) microchannels. Electrophoresis 24(21), 3607–3619 (2003)
F. Meireles, PNA-FISH with microfluidics (Biomedical Engineering, Porto, Faculty of Engineering, 2012)
S.K. Mitra, S. Chakraborty, Microfluidics and nanofluidics handbook: fabrication, implementation, and applications (CRC Press, Boca Raton, 2011)
D. Moreira, Integration of microfluidics and fluorescence In Situ hybridization (FISH) for the rapid identification of microorganisms (Biomedical Engineering, Porto, Faculty of Engineering, 2014)
H. Morgan, D. Holmes, N. G. Green, 3D focusing of nanoparticles in microfluidic channels. IEE Proc. Nanobiotechnol. 150(2), 76–81 (IET, 2003)
A. Moter, U.B. Göbel, Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 41(2), 85–112 (2000)
P.E. Nielsen, Peptide nucleic acid: a versatile tool in genetic diagnostics and molecular biology. Curr. Opin. Biotechnol. 12(1), 16–20 (2001)
P.E. Nielsen, G. Haaima, Peptide nucleic acid (PNA). A DNA mimic with a pseudopeptide backbone. Chem. Soc. Rev. 26(2), 73–78 (1997)
P.E. Nielsen, M. Egholm, R.H. Berg, O. Buchardt, Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 254(5037), 1497–1500 (1991)
S. Patankar, Numerical heat transfer and fluid flow (CRC press, Boca Raton, 1980)
H. Perry-O'Keefe, S. Rigby, K. Oliveira, D. Sørensen, H. Stender, J. Coull, J. Hyldig-Nielsen, Identification of indicator microorganisms using a standardized PNA FISH method. J. Microbiol. Methods 47(3), 281–292 (2001)
C. Probst, A. Grünberger, W. Wiechert, D. Kohlheyer, Polydimethylsiloxane (PDMS) sub-micron traps for single-cell analysis of bacteria. Micromachines 4(4), 357–369 (2013)
A. Rohde, J.A. Hammerl, B. Appel, R. Dieckmann, S. Al Dahouk, FISHing for bacteria in food–a promising tool for the reliable detection of pathogenic bacteria? Food Microbiol. 46, 395–407 (2015)
A.H. Rose, Chemical microbiology: an introduction to microbial physiology (Elsevier, Amsterdam, 2014)
R.S. Santos, N. Guimarães, P. Madureira, N.F. Azevedo, Optimization of a peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) method for the detection of bacteria and disclosure of a formamide effect. J. Biotechnol. 187, 16–24 (2014)
H. Sharma, D. Nguyen, A. Chen, V. Lew, M. Khine, Unconventional low-cost fabrication and patterning techniques for point of care diagnostics. Ann. Biomed. Eng. 39(4), 1313–1327 (2011)
V. Sieben, C.D. Marun, P. Pilarski, G. Kaigala, L. Pilarski, C. Backhouse, FISH and chips: chromosomal analysis on microfluidic platforms. IET Nanobiotechnol. 1(3), 27–35 (2007)
V.J. Sieben, C.S. Debes-Marun, L.M. Pilarski, C.J. Backhouse, An integrated microfluidic chip for chromosome enumeration using fluorescence in situ hybridization. Lab Chip 8(12), 2151–2156 (2008)
H. Stender, M. Fiandaca, J.J. Hyldig-Nielsen, J. Coull, PNA for rapid microbiology. J. Microbiol. Methods 48(1), 1–17 (2002)
J.H. Sung, M.L. Shuler, Prevention of air bubble formation in a microfluidic perfusion cell culture system using a microscale bubble trap. Biomed. Microdevices 11(4), 731–738 (2009)
S. Thorslund, O. Klett, F. Nikolajeff, K. Markides, J. Bergquist, A hybrid poly (dimethylsiloxane) microsystem for on-chip whole blood filtration optimized for steroid screening. Biomed. Microdevices 8(1), 73–79 (2006)
I. Vedarethinam, P. Shah, M. Dimaki, Z. Tumer, N. Tommerup, W.E. Svendsen, Metaphase FISH on a chip: miniaturized microfluidic device for fluorescence in situ hybridization. Sensors 10(11), 9831–9846 (2010)
J.R. Warner, "the economics of ribosome biosynthesis in yeast." trends Biochem. Sci. 24(11), 437–440 (1999)
C. Xi, S.A. Boppart, L. Raskin, Use of molecular beacons for the detection of bacteria in microfluidic devices. Micromachining and Microfabrication, International Society for Optics and Photonics. 4982, 170–177 (2003)
Y. Xia, G.M. Whitesides, Soft lithography. Annu. Rev. Mater. Sci. 28(1), 153–184 (1998)
A. Xufre, H. Albergaria, J. Inácio, I. Spencer-Martins, F. Gírio, Application of fluorescence in situ hybridisation (FISH) to the analysis of yeast population dynamics in winery and laboratory grape must fermentations. Int. J. Food Microbiol. 108(3), 376–384 (2006)
H. Yamazoe, Y. Sugiyama, A. El Omri, Y. Hagihara, T. Okada, Facile immunostaining and labeling of nonadherent cells using a microfluidic device to entrap the cells. J. Biosci. Bioeng. 117(3), 375–378 (2014)
S.F. Yeo, B. Wong, Current status of nonculture methods for diagnosis of invasive fungal infections. Clin. Microbiol. Rev. 15(3), 465–484 (2002)
E.W. Young, D.J. Beebe, Fundamentals of microfluidic cell culture in controlled microenvironments. Chem. Soc. Rev. 39(3), 1036–1048 (2010)
Q. Zhang, L. Zhu, H. Feng, S. Ang, F.S. Chau, W.-T. Liu, Microbial detection in microfluidic devices through dual staining of quantum dots-labeled immunoassay and RNA hybridization. Anal. Chim. Acta 556(1), 171–177 (2006)
Y. Zhang, C. Luo, K. Zou, Z. Xie, O. Brandman, Q. Ouyang, H. Li, Single cell analysis of yeast replicative aging using a new generation of microfluidic device. PLoS One 7(11), e48275 (2012)
Acknowledgements
This work was financially supported by: 1) POCI-01-0145-FEDER-006939 (Laboratory for Process Engineering, Environment, Biotechnology and Energy – UID/EQU/00511/2013) funded by the European Regional Development Fund (ERDF), through COMPETE2020 - Programa Operacional Competitividade e Internacionalização (POCI) and by national funds, through FCT - Fundação para a Ciência e a Tecnologia; 2) NORTE‐01‐0145‐FEDER‐000005 – LEPABE-2-ECO-INNOVATION, supported by North Portugal Regional Operational Programme (NORTE 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (ERDF) and 3) FCT (Scholarship SFRH/BPD/98525/2013) and Project NanoDiaBac (ENMed/0003/2014).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical statements
All authors declare that they have no conflict of interest.
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
André M. Ferreira and Daniela Cruz-Moreira contributed equally to this work.
Rights and permissions
About this article
Cite this article
Ferreira, A.M., Cruz-Moreira, D., Cerqueira, L. et al. Yeasts identification in microfluidic devices using peptide nucleic acid fluorescence in situ hybridization (PNA-FISH). Biomed Microdevices 19, 11 (2017). https://doi.org/10.1007/s10544-017-0150-y
Published:
DOI: https://doi.org/10.1007/s10544-017-0150-y