Abstract
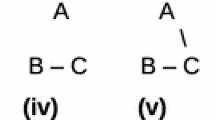
Bayesian methods have become among the most popular methods in phylogenetics, but theoretical opposition to this methodology remains. After providing an introduction to Bayesian theory in this context, I attempt to tackle the problem mentioned most often in the literature: the “problem of the priors”—how to assign prior probabilities to tree hypotheses. I first argue that a recent objection—that an appropriate assignment of priors is impossible—is based on a misunderstanding of what ignorance and bias are. I then consider different methods of assigning prior probabilities to trees. I argue that priors need to be derived from an understanding of how distinct taxa have evolved and that the appropriate evolutionary model is captured by the Yule birth–death process. This process leads to a well-known statistical distribution over trees. Though further modifications may be necessary to model more complex aspects of the branching process, they must be modifications to parameters in an underlying Yule model. Ignoring these Yule priors commits a fallacy leading to mistaken inferences both about the trees themselves and about macroevolutionary processes more generally.



Similar content being viewed by others
References
Aldous DJ (2001) Stochastic models and descriptive statistics for phylogenetic trees, from yule to today. Stat Sci 16(1):23–34
Brown JKM (1994) Probabilities of evolutionary trees. Syst Biol 43(1):78–91
Centers for Disease Control and Prevention (2006) HIV/AIDS Surveillance Report, 2005, vol 17. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, pp 1–54. Also available at http://www.cdc.gov/hiv/topics/surveillance/resources/reports/2005report/
Edwards AWF (1970) Estimation of the branch points of a branching diffusion process. J R Stat Soc B (Methodological) 32(2):155–174
Farris JS (1983) The logical basis of phylogenetic analysis. Adv Cladistics 2:7–36
Felsenstein J (2004) Inferring phylogenies. Sinauer Associates, Sunderland, Mass
Goloboff PA, Pol D (2005) Parsimony and Bayesian phylogenetics. In: Victor AA (ed) Parsimony, phylogeny and genomics. Oxford University Press, Oxford, pp 148–159
Haber MH (2005) On probability and systematics: possibility, probability, and phylogenetic inference. Syst Biol 54(5):831–841
Halliburton R (2004) Introduction to population genetics. Pearson/Prentice Hall, Upper Saddle River
Harding EF (1971) The probabilities of rooted tree-shapes generated by random bifurcation. Adv Appl Probab 3(1):44–77
Hein J, Schierup M, Wiuf C (2005) Gene genealogies, variation and evolution: a primer in coalescent theory. Oxford University Press, Oxford
Howson C, Urbach P (2005) Scientific reasoning: the Bayesian approach, 3rd edn. Open Court, La Salle
Huelsenbeck JP, Kirkpatrick M (1996) Do phylogenetic methods produce trees with biased shapes? Evolution 50(4):1418–1424
Huelsenbeck JP, Ronquist F (2001) MR BAYES: Bayesian inference of phylogenetic trees. Bioinformatics (Oxford) 17(8):754–755
Huelsenbeck JP, Ronquist F (2005) Bayesian analysis of molecular evolution using Mr. Bayes. In: Nielson R (ed) Statistical methods in molecular evolution. Springer, New York, pp 183–232
Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science (Washington DC) 294(5550):2310–2314
Joyce J (2005) How probabilities reflect evidence. Philos Perspect 19:153
Kadane JB (2006) Is “objective Bayesian analysis” objective, Bayesian, or wise? (comment on articles by Berger and by Goldstein). Bayesian Anal 1(3):433–436
Kingman JFC (1982) The coalescent. Stochastic Process Appl 13(3):235–248
Kluge AG (2005) What is the rationale for ‘Ockham’s razor’ (a.k.a. parsimony) in phylogenetic inference? In: Albert VA (ed) Parsimony, phylogeny and genomics. Oxford University Press, Oxford, pp 15–42
Larget B (2005) Introduction to Markov Chain Monte Carlo methods in molecular evolution. In: Nielson R (ed) Statistical methods in molecular evolution. Springer, New York, pp 44–61
Larget B, Simon DL (1999) Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Mol Biol Evol 16(6):750–759
Metzker ML, Mindell DP, Liu XM, Ptak RG, Gibbs RA, Hillis DM (2002) Molecular evidence of HIV-1 transmission in a criminal case. Proc Nat Acad Sci 99(22):14292–14297
Mooers AO, Heard SB (1997) Inferring evolutionary process from phylogenetic tree shape. Q Rev Biol 72(1):31–54
Pickett KM, Randle CP (2005) Strange Bayes indeed: uniform topological priors imply non-uniform clade priors. Mol Phylogenet Evol 34(1):203–211
Pitnick S, Jones KE, Wilkinson GS (2006) Mating system and brain size in bats. Proc R Soc Biol Sci B 273(1587):719–724
Poux C, Madsen O, Marquard E, Vieites DR, de Jong WW, Vences M (2005) Asynchronous colonization of Madagascar by the four endemic clades of primates, tenrecs, carnivores, and rodents as inferred from nuclear genes. Syst Biol 54(5):719–730
Randle CP, Mort ME, Crawford DJ (2005) Bayesian inference of phylogenetics revisited: developments and concerns. Taxon 54(1):9–15
Rannala B, Yang Z (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol 43(3):304–311
Semple C, Steel M (2003). Phylogenetics. Oxford University Press, Oxford
Siddall ME, Kluge AG (1997). Probabilism and phylogenetic inference. Cladistics 13(4):313–336
Sober E (1988). Reconstructing the past. Parsimony, evolution, and inference. MIT Press, Cambridge
Steel M, McKenzie A (2001). Properties of phylogenetic trees generated by yule-type speciation models. Math Biosci 170(1):91–112
Thompson EA (1975) Human evolutionary trees. Cambridge University Press, Cambridge
Velasco JD (2007) Why non-uniform priors on clades are both unavoidable and unobjectionable. Mol Phylogenet Evol 45:748–749
Yang Z (2006) Computational molecular evolution. Oxford University Press, Oxford
Yang Z, Rannala B (1997). Bayesian phylogenetic inference using DNA sequences: a Markov Chain Monte Carlo method. Mol Biol Evol 14(7):717–724
Yule GU (1925) A mathematical theory of evolution, based on the conclusions of Dr. JC Willis, FRS. Philos Trans R Soc Lond B 213:21–87
Acknowledgements
I am grateful to David Baum, Matt Haber, James Justus, Bret Larget, Greg Novack, and especially Elliott Sober for their support and many helpful comments on earlier drafts of this paper. Thanks also to an anonymous reviewer who made several helpful comments which improved the presentation of this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Velasco, J.D. The prior probabilities of phylogenetic trees. Biol Philos 23, 455–473 (2008). https://doi.org/10.1007/s10539-007-9105-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10539-007-9105-7